













| General Information: |
|
| Name(s) found: |
PAH1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 904 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
lipid metabolic process
[IGI]
cellular response to phosphate starvation [IGI] |
| Molecular Function: |
phosphatidate phosphatase activity
[IGI]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLVGRVGSL ISQGVYSVAT PFHPFGGAID VIVVQQQDGS FRSTPWYVRF GKFQGVLKGA 60
61 EKFVRISVNG TEADFHMYLD NSGEAYFIRE VDPAANDTNN LISGSENNNG NQNNGVTYRL 120
121 EHSLSDSGTG ELREGFDPLS RLERTESDCN RRFYDFQDDP PSPTSEYGSA RFDNLNVESY 180
181 GDSQGSDSEV VLVSIDGHIL TAPVSVAEQE AENLRLNTPQ FHLAPGDGTE FCEGNTEFAS 240
241 SETPWDTEYI DKVEESSDTA NIASDKVDAI NDERNDLDSH SRDNAEKDSH DAERDLLGSC 300
301 LEQSELTKTS ENVKSEEPGP TFEDRNLKEG EFPLRTIMEN DRSEDEVTIE SIDTLVDSFE 360
361 SSTTQITIEE VKTTEGSRIS VDSNADSECK DEQTSAETAI LFNNQESSIS VDSNADSECK 420
421 DEQPRISAET AILINNQEGG IIESEDQDSE RVSIDSTREE VDKDNEDRKT VVSVGVTSSV 480
481 DEGEPDTDQR YELSLCKDEL RQGMGLSAAA EVFDAHMISK EEYINSATSI LESENLVVRI 540
541 RETYMPWTKA ARIVLGKAVF DLDLDIQPDD VISVEENESP KPKDDETTIT PSSSGTRWRL 600
601 WPIPFRRVKT VEHTGSNSSS EEDLFVDSEP GLQNSPETQS TTESRHESPR RQLVRTNVPT 660
661 NEQIASLNLK DGQNMITFSF STRVLGTQQV DAHIYRWRWD TKIVISDVDG TITKSDVLGQ 720
721 FMPFIGKDWT QSGVAKLFSA IKENGYQLLF LSARAIVQAY LTRNFLNNLK QDGKALPTGP 780
781 VVISPDGLFP ALYREVIRRA PHEFKIACLE DIRKLFPTDY NPFYAGFGNR DTDELSYRKL 840
841 GIPKGKIFII NPKGEVATGH RIDVKKSYTS LHTLVNDMFP PTSLVEQEDY NPWNFWKLPI 900
901 EEVE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
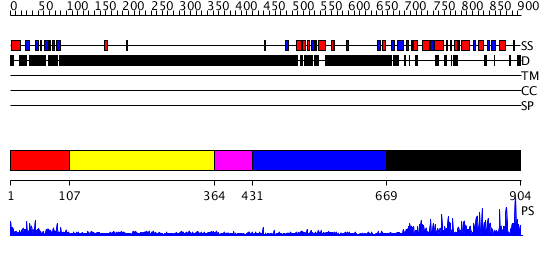
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..106] | 68.657577 | No description for PF04571.6 was found. No confident structure predictions are available. | |
| 2 | View Details | [107..363] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [364..430] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [431..668] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [669..904] | 96.19382 | No description for PF08235.5 was found. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.73 |
Source: Reynolds et al. (2008)