













| General Information: |
|
| Name(s) found: |
NADE_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 725 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
nitrogen compound metabolic process
[IEA][ISS]
NAD biosynthetic process [IEA] |
| Molecular Function: |
hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds
[IEA][ISS]
NAD+ synthase (glutamine-hydrolyzing) activity [IEA] ATP binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRLLKVATCN LNQWAMDFES NMKNIKASIA EAKAAGAVIR LGPELEVTGY GCEDHFLELD 60
61 TVTHAWECLK ELLLGDWTDD ILCSIGMPVI KGAERYNCQV LCMNRRIIMI RPKMWLANDG 120
121 NYRELRWFTA WKQREELEEF QLPIEISEAL EQKSVPFGYG YIQFIDTAVA AEVCEELFSP 180
181 LPPHAELALN GVEVFMNASG SHHQLRKLDI RLNAFMGATH ARGGVYMYSN QQGCDGSRLY 240
241 YDGCACIVVN GNVVAQGSQF SLRDVEVIIS QVDLDAVASL RGSISSFQEQ ASCKVKVSSV 300
301 AVPCRLTQSF NLKMTLSSPK KIIYHSPQEE IAFGPACWMW DYLRRSGASG FLLPLSGGAD 360
361 SSSVAAIVGC MCQLVVKEIA KGDEQVKADA NRIGNYANGQ FPTDSKEFAK RIFYTVFMGS 420
421 ENSSEETKRR SKQLADEIGA WHLDVCIDGV VSAVLSLFQT VTGKRPRYKV DGGSNAENLG 480
481 LQNIQARMRM VLAFMLASLL PWVHSKPGFY LVLGSSNVDE GLRGYLTKYD CSSADINPIG 540
541 SISKMDLRLF LKWAATNLGY PSLAEIEAAP PTAELEPIRS DYSQLDEVDM GMTYEELSVY 600
601 GRMRKIFRCG PVSMFKNLCY KWGTKLSPAE VAEKVKYFFK YYSINRHKMT VLTPSYHAES 660
661 YSPEDNRFDL RQFLYNSKWP YQFKKIDEIV DSLNGDSVAF PEEEANSNKE IGVVAANSGD 720
721 PSAGL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
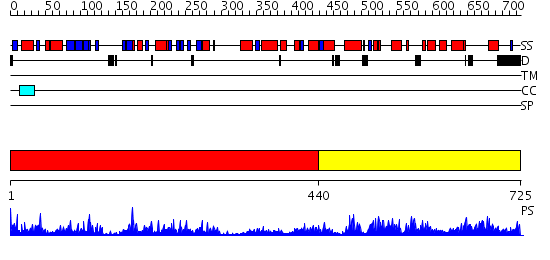
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..439] | 60.69897 | NIT-FHIT fusion protein, C-terminal domain; NIT-FHIT fusion protein, N-terminal domain | |
| 2 | View Details | [440..725] | 57.69897 | E.coli NAD Synthetase, AMP |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 |
|
||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)