













| General Information: |
|
| Name(s) found: |
RGAP6_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 827 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[IDA]
|
| Biological Process: |
signal transduction
[ISS]
|
| Molecular Function: |
phosphoinositide binding
[ISS]
Rho GTPase activator activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEASLAVIQR PQAGASNTVY KSGPLFISSK GLGWTSWKKR WFILTRTSLV FFKNDPSALP 60
61 QKGGEVNLTL GGIDLNSSGS VVVREDKKLL TVLFPDGRDG RAFTLKAETL DDLYEWKAAL 120
121 EQALAQAPNA ALVIGQNGIF RTEANNTIEA SFNSWRDQRP LKSSVVGRPI LLALEEIDGS 180
181 PSFLEKALQF LETYGTKVEG ILRQSADVEE VERRVQEYEQ GKTEFSPEED PHVVGDCVKH 240
241 VLRQLPSSPV PASCCTALLE AYKIDQNEAR VNSLRSAIIE TFPEPNRRLL LRMLKMMHTI 300
301 TSHSSENRMT SSAVAACMSP LLLRPLLAGE CDLEGFDTLG DNSAQLLAAA NAANNAQAIV 360
361 TALLEDYGNM INDEGLGRCS TSTDSHIGDS GPENSSDEEE IVVKHPDLHT LDIEEGETDD 420
421 DNDVLLSRKP SESSDYAGSD LYDYKGFGVE DSDAESPRDI HCSVESTDFS ARVKKHIEEP 480
481 IKDIEVSSVS PTENCYQSGR EAIPSVTPST PLTALRYTTS AEKPASKTTG SSTVNSKRSS 540
541 SWGRGNGKKT PAKGSFDSSG NDELLIQRLE HMKDELRQRI AKEAKGNAAL QASLERRKQA 600
601 LHERRLALEQ DVGRLQEQLQ AERDLRSALE VGLSISCGQF SSQAADSKTR AELEEIALAE 660
661 ADVARLKQKV AELHHQLSQQ RQHHLSSLPD AQSHHQFLHN HNTQLKSFQQ DFDSILAFVN 720
721 HERNQRTDET SLRADWRNGR GNNRQVPGSP SLNAASLGIP MEEYSPVMDY GRHHHPPATS 780
781 AALMELTTRL DFFKERRSQL MEQIQNLDLN YGSSSSSLHR SSSPPWN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
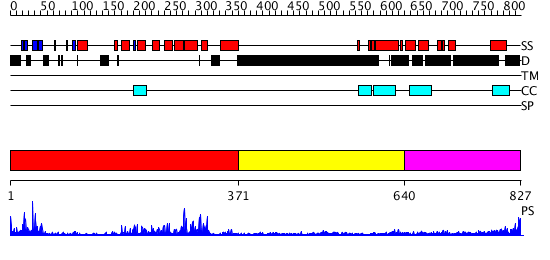
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..370] | 47.69897 | Crystal Structure of the Human Beta2-Chimaerin | |
| 2 | View Details | [371..639] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [640..827] | 2.26499 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)