













| General Information: |
|
| Name(s) found: |
gi|9755633
[NCBI NR]
gi|15237207 [NCBI NR] gi|11357570 [NCBI NR] |
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 745 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: |
endo-1,3(4)-beta-glucanase activity
[IEA]
hydrolase activity, acting on glycosyl bonds [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLKKVRRKVK VLITKPFKKP KNRPPSPPPP LPLPPSPSPP PSQQMSSSRQ KNTPFLFPRS 60
61 DSSVLPDPSR FFSHDLLSSP LPTNSFFQNF TLNNGDQAEY FHPYIIKPST SSLSISYPSL 120
121 SHNSAFIYEA FNADITITGS DGPDPHSRKS HLISSFSDLG VTLDFPSSNL RFFLVRGSPF 180
181 ITFSVNSSIT ISTIHAVLSL SGNTSSTKYT VKLNNNQTWL IYASSPINLT KDGVSSINCG 240
241 DGFSGIIRIV VLPNPNPYFE TILDGFSCSY PVSGDADFTK PFALEYKWEK RGYGDLLMLA 300
301 HPLHLKLLST NDCSITVLDN FKYNSIDGDL VGVIGDSWVL KPDPVSVTWH SIKGVQEDSH 360
361 QEIISALIKD VNALDSSAEV TNSSYFYAKL IARAARLALI AEEVCYLDVI PKIRTYLKNM 420
421 IEPWLNGSFG PNGFLYDPKW GGVITKLGSR DSGADFGFGI YNDHHYHLGY FVYAIAVLAK 480
481 IDPLWGKRYR PQAYTLMADY LTLGKKGAKS NSNYPRLRCF DLFKLHSWAG GLTEFADGRN 540
541 QESTSEAVNA YYSAALLGLA YGDTHLVAAA SMVLTLEIHA AKMWWQVKED DAIYPQDFTS 600
601 ENRVVGVLWS TKRDSGLWFA PKEWKECRLG IQLLPLLPVS EVLFSDVTFV KQLVNWTMPA 660
661 LARDSVGEGW KGFVYALESM YDKDGAMEKI KGLNGFDDGN SLSNLLWWVH SRNNDDDDDY 720
721 EEDDEGGYGG HGGGGKYCSF GHYCH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
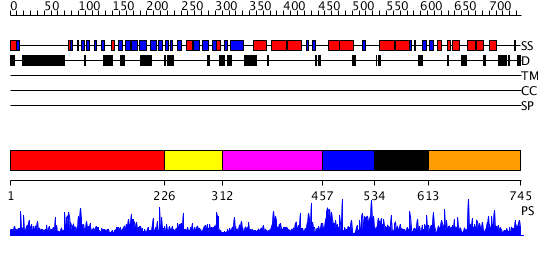
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..225] | 1.041988 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [226..311] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [312..456] | 1.044982 | View MSA. Confident ab initio structure predictions are available. | |
| 4 | View Details | [457..533] | N/A | Confident ab initio structure predictions are available. | |
| 5 | View Details | [534..612] | 1.044991 | View MSA. Confident ab initio structure predictions are available. | |
| 6 | View Details | [613..745] | 2.043976 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.55 |
Source: Reynolds et al. (2008)