













| General Information: |
|
| Name(s) found: |
gi|9759100
[NCBI NR]
gi|20258919 [NCBI NR] gi|15810415 [NCBI NR] gi|15239227 [NCBI NR] |
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 294 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
protein folding
[IEA][ISS]
|
| Molecular Function: |
heat shock protein binding
[IEA]
unfolded protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDGNKDDALK CLKIGKDAIE AGDRSRALKF LEKARRLDPN LPIDGLVSDL KKQSDEPAAE 60
61 EDSPGSAANE SSKPSDRPSL RQRGSSSSAA GSSSSSSSTE EQRTIVREIK SKKDYYEILG 120
121 LKSNCSVEDL RKSYRKLSLK VHPDKNKAPG SEEAFKSVSK AFQCLSNEDT RRKYDGSGSD 180
181 EPAYQPRRDA RRNNGFNGFY DDEFDADEIF RSFFGGGEMN PATTQFRSFN FGGGTRTANQ 240
241 ASDTGFNPRV LLQILPVVFI LLLNFLPSPQ PIYSLSHRIT TSSNSPRIGA SLTL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
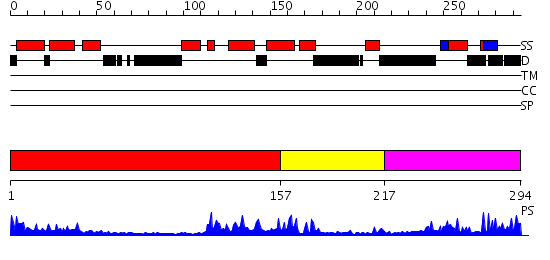
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..156] | 3.30103 | Crystal structure of an 8 repeat consensus TPR superhelix (trigonal crystal form) | |
| 2 | View Details | [157..216] | 23.0 | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | |
| 3 | View Details | [217..294] | 11.522879 | The crystal structure of Hsp40 Ydj1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 |
|
|||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.47 |
Source: Reynolds et al. (2008)