













| General Information: |
|
| Name(s) found: |
SODF2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 305 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleoid
[IDA]
chloroplast [IDA] |
| Biological Process: |
response to UV
[IEP]
|
| Molecular Function: |
superoxide dismutase activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMNVAVTATP SSLLYSPLLL PSQGPNRRMQ WKRNGKRRLG TKVAVSGVIT AGFELKPPPY 60
61 PLDALEPHMS RETLDYHWGK HHKTYVENLN KQILGTDLDA LSLEEVVLLS YNKGNMLPAF 120
121 NNAAQAWNHE FFWESIQPGG GGKPTGELLR LIERDFGSFE EFLERFKSAA ASNFGSGWTW 180
181 LAYKANRLDV ANAVNPLPKE EDKKLVIVKT PNAVNPLVWD YSPLLTIDTW EHAYYLDFEN 240
241 RRAEYINTFM EKLVSWETVS TRLESAIARA VQREQEGTET EDEENPDDEV PEVYLDSDID 300
301 VSEVD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
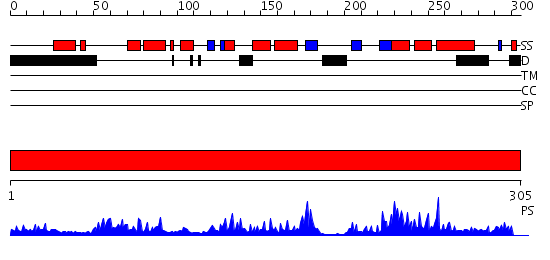
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..305] | 75.0 | The crystal structure of the eukaryotic FeSOD from Vigna unguiculata suggests a new enzymatic mechanism |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.47 |
Source: Reynolds et al. (2008)