













| General Information: |
|
| Name(s) found: |
TPS10_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 861 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
metabolic process
[IEA]
trehalose biosynthetic process [IEA][ISS] |
| Molecular Function: |
transferase activity, transferring glycosyl groups
[ISS]
trehalose-phosphatase activity [IMP][ISS] [NOT]alpha,alpha-trehalose-phosphate synthase (UDP-forming) activity [IMP] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGSKSFGNLL DLASGDLLDI PQTPRYLPRV MTVPGIISDV DGYGISDGDS DVISLPCRER 60
61 KIIVANFLPL NGKKDSETGK WKFSLDNDSP LLHLKDGFSP ETEVIYVGSL KTHVDVSEQD 120
121 EVSHNLFEEF NCVATFLPQD VHKKFYLGFC KQQLWPLFHY MLPMCPDHGE RFDRGLWQAY 180
181 VSANKIFADK VMGVINLEED YIWIHDYHLM VLPTFLRRRF HRVKLGFFLH SPFPSSEIYR 240
241 TLPVREELLR GLLNCDLIGF HTFDYARHFL SCCCRMLGLE YESKRGHIAL DYLGRTVFLK 300
301 ILPIGIHMGR LESVLNLPAT AEKLKEIQEK YRGKKIILGV DDMDIFKGLS LKILAFEHLL 360
361 QQYPSMLGKI VLIQIVNPAR GSGKDVQEAR KETYDTVKRI NERYGSHDYE PVVLIDRPVP 420
421 RFEKSAYYAL AECCIVNAVR DGMNLVPYKY TVCRQGTPSM NKSLGVSDDL PRTSTLVLSE 480
481 FIGCSPSLSG AIRVNPWDVD AVADSLYSAI TMSDFEKQLR HKKHFHYIST HDVGYWARSF 540
541 SQDLERASRD HYSKRCWGVG WGLGFRLVAL SPNFRRLSIE QTVSAYRRSS KRAIFLDYDG 600
601 TLVPETSIVK DPSAEVISAL KALCSDPNNT IFIVSGRGKV SLSEWLAPCE NLGIAAEHGY 660
661 FTRWNKSSDW ETSGLSDDLE WKKVVEPIMR LYTETTDGSN IEAKESALVW HHQDADPDFG 720
721 SCQAKELLDH LETVLVNEPV IVNRGHQIVE VKPQGVSKGL VTGKILSRML EDGIAPDFVV 780
781 CIGDDRSDEE MFENISTTLS AQSSSMSTEI FACTVGRKPS KAKYFLDEVS DVVKLLQGLA 840
841 NTSSPKPRYP SHLRVSFESV V |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
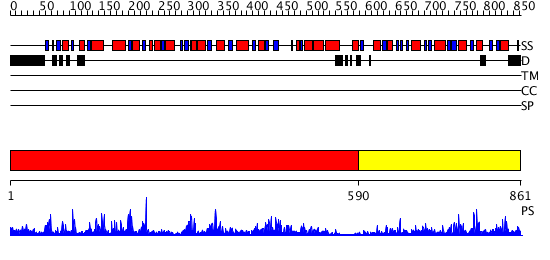
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..589] | 85.522879 | Trehalose-6-phosphate from E. coli bound with UDP-2-fluoro glucose. | |
| 2 | View Details | [590..861] | 26.09691 | Crystal structure of NYSGRC target T1436: A Hypothetical protein yidA. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)