













| General Information: |
|
| Name(s) found: |
ATB14_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 852 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][ISS]
|
| Biological Process: |
integument development
[IGI]
primary shoot apical meristem specification [IMP] regulation of transcription, DNA-dependent [ISS] polarity specification of adaxial/abaxial axis [IMP] adaxial/abaxial pattern formation [IMP] determination of bilateral symmetry [IMP] meristem initiation [IMP] |
| Molecular Function: |
DNA binding
[ISS]
transcription factor activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMMVHSMSRD MMNRESPDKG LDSGKYVRYT PEQVEALERV YTECPKPSSL RRQQLIRECP 60
61 ILSNIEPKQI KVWFQNRRCR EKQRKEAARL QTVNRKLNAM NKLLMEENDR LQKQVSNLVY 120
121 ENGHMKHQLH TASGTTTDNS CESVVVSGQQ HQQQNPNPQH QQRDANNPAG LLSIAEEALA 180
181 EFLSKATGTA VDWVQMIGMK PGPDSIGIVA ISRNCSGIAA RACGLVSLEP MKVAEILKDR 240
241 PSWLRDCRSV DTLSVIPAGN GGTIELIYTQ MYAPTTLAAA RDFWTLRYST CLEDGSYVVC 300
301 ERSLTSATGG PTGPPSSNFV RAEMKPSGFL IRPCDGGGSI LHIVDHVDLD AWSVPEVMRP 360
361 LYESSKILAQ KMTVAALRHV RQIAQETSGE VQYGGGRQPA VLRTFSQRLC RGFNDAVNGF 420
421 VDDGWSPMGS DGAEDVTVMI NLSPGKFGGS QYGNSFLPSF GSGVLCAKAS MLLQNVPPAV 480
481 LVRFLREHRS EWADYGVDAY AAASLRASPF AVPCARAGGF PSNQVILPLA QTVEHEESLE 540
541 VVRLEGHAYS PEDMGLARDM YLLQLCSGVD ENVVGGCAQL VFAPIDESFA DDAPLLPSGF 600
601 RIIPLEQKST PNGASANRTL DLASALEGST RQAGEADPNG CNFRSVLTIA FQFTFDNHSR 660
661 DSVASMARQY VRSIVGSIQR VALAIAPRPG SNISPISVPT SPEALTLVRW ISRSYSLHTG 720
721 ADLFGSDSQT SGDTLLHQLW NHSDAILCCS LKTNASPVFT FANQTGLDML ETTLVALQDI 780
781 MLDKTLDEPG RKALCSEFPK IMQQGYAHLP AGVCASSMGR MVSYEQATVW KVLEDDESNH 840
841 CLAFMFVNWS FV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
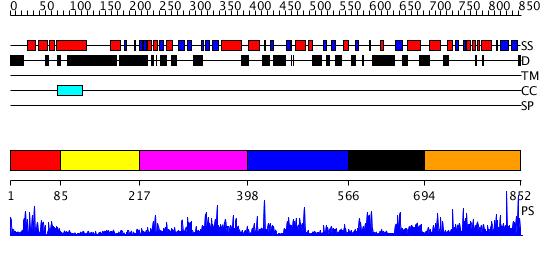
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..84] | 16.69897 | Homeobox protein hox-b1 | |
| 2 | View Details | [85..216] | 2.033996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [217..397] | 2.084979 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [398..565] | 1.05998 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [566..693] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [694..852] | 93.309804 | No description for PF08670.3 was found. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)