













| General Information: |
|
| Name(s) found: |
REV_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 842 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][ISS]
|
| Biological Process: |
xylem and phloem pattern formation
[IMP]
cell differentiation [IMP] regulation of transcription, DNA-dependent [ISS] radial pattern formation [IMP] polarity specification of adaxial/abaxial axis [IMP] determination of bilateral symmetry [IMP] meristem initiation [IMP] |
| Molecular Function: |
DNA binding
[ISS]
transcription factor activity [ISS] lipid binding [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEMAVANHRE RSSDSMNRHL DSSGKYVRYT AEQVEALERV YAECPKPSSL RRQQLIRECS 60
61 ILANIEPKQI KVWFQNRRCR DKQRKEASRL QSVNRKLSAM NKLLMEENDR LQKQVSQLVC 120
121 ENGYMKQQLT TVVNDPSCES VVTTPQHSLR DANSPAGLLS IAEETLAEFL SKATGTAVDW 180
181 VQMPGMKPGP DSVGIFAISQ RCNGVAARAC GLVSLEPMKI AEILKDRPSW FRDCRSLEVF 240
241 TMFPAGNGGT IELVYMQTYA PTTLAPARDF WTLRYTTSLD NGSFVVCERS LSGSGAGPNA 300
301 ASASQFVRAE MLSSGYLIRP CDGGGSIIHI VDHLNLEAWS VPDVLRPLYE SSKVVAQKMT 360
361 ISALRYIRQL AQESNGEVVY GLGRQPAVLR TFSQRLSRGF NDAVNGFGDD GWSTMHCDGA 420
421 EDIIVAINST KHLNNISNSL SFLGGVLCAK ASMLLQNVPP AVLIRFLREH RSEWADFNVD 480
481 AYSAATLKAG SFAYPGMRPT RFTGSQIIMP LGHTIEHEEM LEVVRLEGHS LAQEDAFMSR 540
541 DVHLLQICTG IDENAVGACS ELIFAPINEM FPDDAPLVPS GFRVIPVDAK TGDVQDLLTA 600
601 NHRTLDLTSS LEVGPSPENA SGNSFSSSSS RCILTIAFQF PFENNLQENV AGMACQYVRS 660
661 VISSVQRVAM AISPSGISPS LGSKLSPGSP EAVTLAQWIS QSYSHHLGSE LLTIDSLGSD 720
721 DSVLKLLWDH QDAILCCSLK PQPVFMFANQ AGLDMLETTL VALQDITLEK IFDESGRKAI 780
781 CSDFAKLMQQ GFACLPSGIC VSTMGRHVSY EQAVAWKVFA ASEENNNNLH CLAFSFVNWS 840
841 FV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
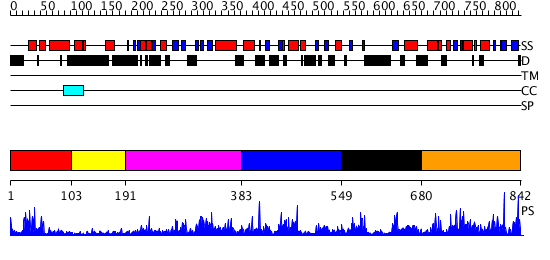
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..102] | 12.39794 | Homeobox protein hox-b1 | |
| 2 | View Details | [103..190] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [191..382] | 27.30103 | No description for 2psoA was found. | |
| 4 | View Details | [383..548] | 1.068979 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [549..679] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [680..842] | 90.677781 | No description for PF08670.3 was found. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)