













| General Information: |
|
| Name(s) found: |
gi|62319772
[NCBI NR]
gi|51970450 [NCBI NR] gi|51970270 [NCBI NR] gi|51969540 [NCBI NR] gi|51969292 [NCBI NR] gi|26451129 [NCBI NR] gi|22137018 [NCBI NR] gi|20197735 [NCBI NR] gi|18401727 [NCBI NR] gi|15983791 [NCBI NR] |
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 607 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IEA]
|
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATSDSRSSP SSSDTEFADP NPSSDPETNS ERVQSQLESM NLSQPSEVSD GSHTEFSGGG 60
61 DDNDDEVASA NGNEGGVSNG GLLREGVAGT SGGEVLLRAE NPVEMEAGEE PPSPTSSGYD 120
121 GERGSSGGAT STYKADDGSE DEIREANVDG DTASQHEAAW LPGKRHVDED DASTSWRKRK 180
181 KHFFILSNSG KPIYSRYGDE HKLAGFSATL QAIISFVENG GDRVNLVKAG NHQVVFLVKG 240
241 PIYLVCISCT DETYEYLRGQ LDLLYGQMIL ILTKSIDRCF EKNAKFDMTP LLGGTDAVFS 300
301 SLVHSFSWNP ATFLHAYTCL PLPYALRQAT GTILQEVCAS GVLFSLLMCR HKVVSLAGAQ 360
361 KASLHPDDLL LLSNFVMSSE SFRTSESFSP ICLPRYNAQA FLHAYVHFFD DDTYVILLTT 420
421 RSDAFHHLKD CRVRLEAVLL KSNILSVVQR SIAEGGMRVE DVPIDRRRRS STTNQEQDSP 480
481 GPDISVGTGG PFGLWHFMYR SIYLDQYISS EFSPPVTSHR QQKSLYRAYQ KLYASMHVKG 540
541 LGPHKTQYRR DENYTLLCWV TPDFELYAAF DPLADKAMAI KICNQVCQRV KDVENEVFLQ 600
601 GASPFSW |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
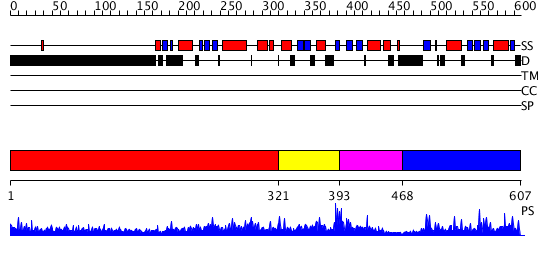
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..320] | 11.045757 | NMR STRUCTURE OF THE TUMOR SUPPRESSOR BIN1: ALTERNATIVE SPLICING IN MELANOMA AND INTERACTION WITH C-MYC | |
| 2 | View Details | [321..392] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [393..467] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [468..607] | N/A | Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)