













| General Information: |
|
| Name(s) found: |
BPM3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 408 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
cytosol [IDA] |
| Biological Process: |
cellular response to salt stress
[IEP]
response to osmotic stress [IEP] |
| Molecular Function: |
protein binding
[IPI][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTVGGIEQL IPDSVSTSFI ETVNGSHQFT IQGYSLAKGM SPGKFIQSDI FSVGGYDWAI 60
61 YFYPDGKNPE DQSSYISLFI ALASDSNDIR ALFELTLMDQ SGKGKHKVHS HFDRALEGGP 120
121 YTLKYKGSMW GYKRFFKRSA LETSDYLKDD CLVINCTVGV VRARLEGPKQ YGIVLPLSNM 180
181 GQGLKDLLDS EVGCDIAFQV GDETYKAHKL ILAARSPVFR AQFFGPIGNN NVDRIVIDDI 240
241 EPSIFKAMLS FIYTDVLPNV HEITGSTSAS SFTNMIQHLL AAADLYDLAR LKILCEVLLC 300
301 EKLDVDNVAT TLALAEQHQF LQLKAFCLEF VASPANLGAV MKSEGFKHLK QSCPTLLSEL 360
361 LNTVAAADKS STSGQSNKKR SASSVLGCDT TNVRQLRRRT RKEVRAVS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
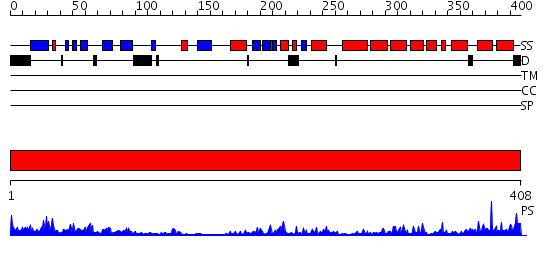
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..408] | 13.39794 | Crystal structure of HAUSP |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)