













| General Information: |
|
| Name(s) found: |
P4KG7_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 650 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: | NONE FOUND |
| Molecular Function: |
inositol or phosphatidylinositol kinase activity
[IDA][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSRNLDSPVQ TQMAVAVFKT PLTGASKMEG KQHHKHQHLQ RQSSGRRVFV QTETGCVLGM 60
61 ELDRSDNVHT VKRRLQIALN FPTEESSLTY GDMVLTNDLS AVRNDSPLLL KRNFMHRSSS 120
121 TPCLSPTGRD LQQKDRSGPI EILGHSDCFS IVKHMVKDIV KAMKMGVEPL PVHSGLGGAY 180
181 YFRNKRGESV AIVKPTDEEP FAPNNPKGFV GKALGQPGLK SSVRVGETGF REVAAYLLDY 240
241 GRFANVPPTA LVKITHSVFN VNDGVKGNKP REKKLVSKIA SFQKFVAHDF DASDHGTSSF 300
301 PVTSVHRIGI LDIRIFNTDR HGGNLLVKKL DGVGMFGQVE LIPIDHGLCL PETLEDPYFE 360
361 WIHWPQASLP FSDEEVDYIQ SLDPVKDCDM LRRELPMIRE ACLRVLVLCT IFLKEASAYG 420
421 LCLAEIGEMM TREFRPGEEE PSELEVVCIE AKRSVTERDV FSPRSDVVGE AEFQFDLDCD 480
481 DLESVYSSKI QLTDDYFTKN PFSNGRSSLG KLEESIKEEE EDEEEEEDKT ENTVPMIIMK 540
541 DSFFSSAAFH DKAPSLSKLS TSMKNTHLSD TTRKNPKPLT RGKSENTSSG HKSANEQLPV 600
601 SASFVKVADM KEDEWVLFLE RFQELLGPAF AKRKTATLSK RQRLGTSCQF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
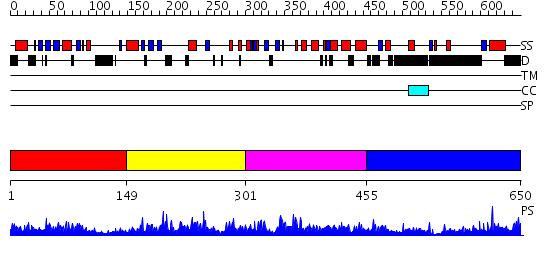
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..148] | 25.0 | Solution Structure of S5a UIM-1/Ubiquitin Complex | |
| 2 | View Details | [149..300] | 2.091968 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [301..454] | 6.07797 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [455..650] | 1.024928 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)