













| General Information: |
|
| Name(s) found: |
MBD13_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 746 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
methyl-CpG binding
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNGEGISDGL SAERKVEIRV RKNGRKDKVI VEKSAAQGLP EGWIKKLEIT NRSGRKTRRD 60
61 PFFIDPKSEY IFQSFKDASR YVETGNIGHY ARKLKESDIE DDDSGNGKTV LRLEYVDKRS 120
121 ADDVLEKEKT IDDVRRSKRR NLSSSDEHSK NCKMTSDLSI VTSQVLEDLG KKEEVKDPIE 180
181 KQLIAKRVTR SQTKASTTEE VVVDLKRNLS SSNAKSEKDS VNSSVRSQKP KKEAVMKEEE 240
241 EQDSSEKRIT RSKVEEKKNE LSNSVARRTS KRLAGIELEP TPELKTRAKV QRIVPLDDEP 300
301 TPELKTRTKV QRVVPPDDEP TPELKTRTKI QRIVPPDDEP TLELKTRTKV QRILPPDDEL 360
361 TPELKSRTKV QRIVPPDDEL TPEFKTRTKV QQRIPPDDGR AGKCKQPVNH VTTSGSKKTE 420
421 IPLNKEVAQS CNEQSSQKPH AAAATSNNRV SADSAVGIQN IGKAVGRKPS KDKKTLKSPL 480
481 IVYELNPVFH LDGYKQKEEM SPVSPLSCQT SATKCEKTAA GKRVGRSSPK ANLTTSVKPT 540
541 QISPLRSPNK GKQPHPSDSG SAIQRRNKLA NEYSNSSVVR GTCSEVMEKS TNSFSSAFDS 600
601 TLADLCKDPC IAFAIKTLTG ESLCLLNTPA ISSNPINNHT KQKGVSFTPE TPGNVNTCSE 660
661 KLVFPSPPPG ANIWQDPCID FAIKTLTGAI PIGLDEPDTK SKSQGMTSTT AATQEAKGRQ 720
721 NNCDYMTNKT VGKPDDLRFT QSFSKD |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
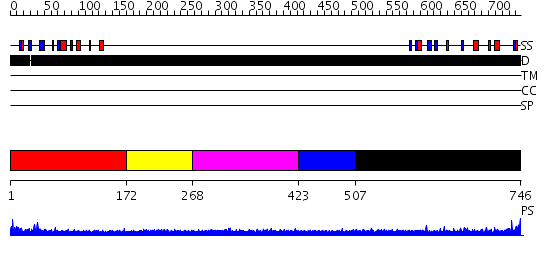
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..171] | 2.164996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [172..267] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [268..422] | 2.185997 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [423..506] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [507..746] | 1.178995 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)