













| General Information: |
|
| Name(s) found: |
Y1301_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 634 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
pollen tube growth
[IEP]
response to light stimulus [ISS] |
| Molecular Function: |
protein binding
[IEA]
signal transducer activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGLVTVGELK PAFTGKRGFR LNSTIRHASE WPISDVSSDL TVQVGSSSFC LHKFPLVSRS 60
61 GKIRKLLADP KISNVCLSNA PGGSEAFELA AKFCYGINIE INLLNIAKLR CASHYLEMTE 120
121 DFSEENLASK TEHFLKETIF PSILNSIIVL HHCETLIPVS EDLNLVNRLI IAVANNACKE 180
181 QLTSGLLKLD YSFSGTNIEP QTPLDWWGKS LAVLNLDFFQ RVISAVKSKG LIQDVISKIL 240
241 ISYTNKSLQG LIVRDPKLEK ERVLDSEGKK KQRLIVETIV RLLPTQGRRS SVPMAFLSSL 300
301 LKMVIATSSS ASTGSCRSDL ERRIGLQLDQ AILEDVLIPI NLNGTNNTMY DIDSILRIFS 360
361 IFLNLDEDDE EEEHHHLQFR DETEMIYDFD SPGSPKQSSI LKVSKLMDNY LAEIAMDPNL 420
421 TTSKFIALAE LLPDHARIIS DGLYRAVDIY LKVHPNIKDS ERYRLCKTID SQKLSQEACS 480
481 HAAQNERLPV QMAVQVLYFE QIRLRNAMSS SIGPTQFLFN SNCHQFPQRS GSGAGSGAIS 540
541 PRDNYASVRR ENRELKLEVA RMRMRLTDLE KDHISIKQEL VKSNPGTKLF KSFAKKISKL 600
601 NSLFSFSSLK PSLSGKASSE SRFLFQRKRR HSVS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
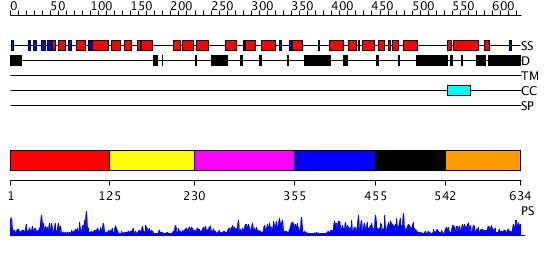
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..124] | 20.522879 | Crystal Structure of the B-Cell Lymphoma 6 (BCL6) BTB domain to 2.2 Angstrom | |
| 2 | View Details | [125..229] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [230..354] | 2.392985 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [355..454] | N/A | Confident ab initio structure predictions are available. | |
| 5 | View Details | [455..541] | 3.073984 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [542..634] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)