













| General Information: |
|
| Name(s) found: |
GTE4_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 766 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
root development
[IMP]
DNA mediated transformation [IMP] cell cycle [IMP] |
| Molecular Function: |
DNA binding
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASEPVNGGE GIDGAREKQI IKVYKRKGKG QRKQSSFFAL EAAIEKPEGL LENENDNNDV 60
61 SPAETLAPEF EDPIVVVKNS IEEAALGTNS HGDKNLTEAP SENLPGDDSD KVIDKPLVEA 120
121 FSQAQPQDDA SLAAMDKSEE VPSQIPKAQD DVNTVVVDEN SIKEPPKSLA QEDVTTVIVD 180
181 KNPIEAPSQT LSLEDGDTLV VDKNPIEVSS EEDVHVIDAD NLIKEAHPEN FVERDTTDAQ 240
241 QPAGLTSDSA HATAAGSMPM EEDADGRIRI HVASTTKQQK EEIRKKLEDQ LNVVRGMVKK 300
301 IEDKEGEIGA YNDSRVLINT GINNGGGRIL SGFASAGLPR EVIRAPRPVN QLSISVLENT 360
361 QGVNEHVEKE KRTPKANQFY RNSEFLLGDK LPPAESNKKS KSSSKKQGGD VGHGFGAGTK 420
421 VFKNCSALLE RLMKHKHGWV FNAPVDVKGL GLLDYYTIIE HPMDLGTIKS ALMKNLYKSP 480
481 REFAEDVRLT FHNAMTYNPE GQDVHLMAVT LLQIFEERWA VIEADYNREM RFVTGYEMNL 540
541 PTPTMRSRLG PTMPPPPINV RNTIDRADWS NRQPTTTPGR TPTSATPSGR TPALKKPKAN 600
601 EPNKRDMTYE EKQKLSGHLQ NLPPDKLDAI VQIVNKRNTA VKLRDEEIEV DIDSVDPETL 660
661 WELDRFVTNY KKGLSKKKRK AELAIQARAE AERNSQQQMA PAPAAHEFSR EGGNTAKKTL 720
721 PTPLPSQVEK QNNETSRSSS SSSSSSSSSS SDSDSDSSSS SGSDQT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
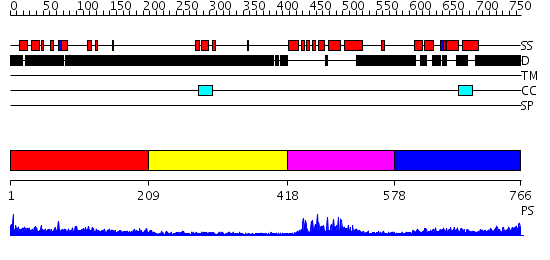
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..208] | 1.188994 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [209..417] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [418..577] | 37.69897 | No description for 2dvvA was found. | |
| 4 | View Details | [578..766] | 2.195991 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)