













| General Information: |
|
| Name(s) found: |
IF4G2_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 747 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
RNA metabolic process
[IEA]
translation [ISS] |
| Molecular Function: |
protein binding
[IEA]
RNA binding [ISS] binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQQQGEPSVL SLRPGGGGGK SRLFVPRFSS SSSFDLTNGG SEETPFPVKR ENSGERVRFS 60
61 REEILQHRES VQVSDEILRR CKEIAVELFG EEQSWGNHAA ESKITNHTQH RHTETDNRDW 120
121 HSRSQIPTSG KEWLRDDPRE AKSTWQGSGP TPVLIKAEVP WSAKRGALSD KDRVVKSVKG 180
181 ILNKLTPEKY ELLKGQLIDA GITSADILKE VIQLIFENAI LQPTFCEMYA LLCFDINGQL 240
241 PSFPSEEPGG KEITFKRVLL NNCQEAFEGA GKLKEEIRQM TNPDQEMERM DKEKMAKLRT 300
301 LGNIRLIGEL LKQKMVPEKI VHHIVQELLG DDTKACPAEG DVEALCQFFI TIGKQLDDSP 360
361 RSRGINDTYF GRLKELARHP QLELRLRFMV QNVVDLRANK WVPRREEVKA KKINEIHSEA 420
421 ERNLGMRPGA MASMRNNNNN RAAVSGAADG MGLGNILGRP GTGGMMPGMP GTRVMPMDED 480
481 GWEMARTRSM PRGNRQTVQQ PRFQPPPAIN KSLSVNSRLL PQGSGGLLNG GGRPSPLLQG 540
541 NGSSSAPQAS KPIPTVEKPQ PRSQPQPQPQ AAPLANSLNA GELERKTKSL LEEYFSIRLV 600
601 DEALQCVEEL KSPSYHPELV KETISLGLEK NPPLVEPIAK LLKHLISKNV LTSKDLGAGC 660
661 LLYGSMLDDI GIDLPKAPNS FGEFLGELVS AKVLDFELVR DVLKKMEDEW FRKTVLNAVI 720
721 KSVRECPSGQ SVLDSQAVEV EACQSLL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
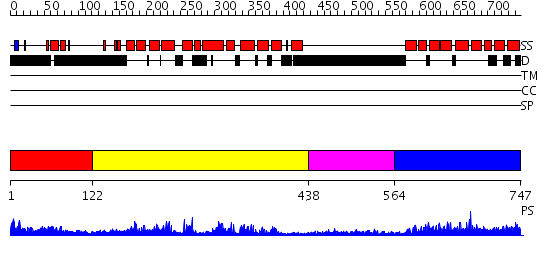
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..121] | 1.164982 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [122..437] | 59.39794 | Eukaryotic initiation factor eIF4G | |
| 3 | View Details | [438..563] | 1.08 | No description for 2j63A was found. | |
| 4 | View Details | [564..747] | 42.0 | C-terminal portion of human eIF4GI |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 |
|
||||||
| 3 | No functions predicted. | ||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)