













| General Information: |
|
| Name(s) found: |
ARR1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 690 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
regulation of anthocyanin metabolic process
[IGI]
shoot development [IGI] primary root development [IGI] regulation of chlorophyll biosynthetic process [IGI] cytokinin mediated signaling pathway [IGI][TAS] response to cytokinin stimulus [IGI][IMP] |
| Molecular Function: |
transcription factor activity
[IDA][ISS]
two-component response regulator activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMNPSHGRGL GSAGGSSSGR NQGGGGETVV EMFPSGLRVL VVDDDPTCLM ILERMLRTCL 60
61 YEVTKCNRAE MALSLLRKNK HGFDIVISDV HMPDMDGFKL LEHVGLEMDL PVIMMSADDS 120
121 KSVVLKGVTH GAVDYLIKPV RMEALKNIWQ HVVRKRRSEW SVPEHSGSIE ETGERQQQQH 180
181 RGGGGGAAVS GGEDAVDDNS SSVNEGNNWR SSSRKRKDEE GEEQGDDKDE DASNLKKPRV 240
241 VWSVELHQQF VAAVNQLGVE KAVPKKILEL MNVPGLTREN VASHLQKYRI YLRRLGGVSQ 300
301 HQGNLNNSFM TGQDASFGPL STLNGFDLQA LAVTGQLPAQ SLAQLQAAGL GRPAMVSKSG 360
361 LPVSSIVDER SIFSFDNTKT RFGEGLGHHG QQPQQQPQMN LLHGVPTGLQ QQLPMGNRMS 420
421 IQQQIAAVRA GNSVQNNGML MPLAGQQSLP RGPPPMLTSS QSSIRQPMLS NRISERSGFS 480
481 GRNNIPESSR VLPTSYTNLT TQHSSSSMPY NNFQPELPVN SFPLASAPGI SVPVRKATSY 540
541 QEEVNSSEAG FTTPSYDMFT TRQNDWDLRN IGIAFDSHQD SESAAFSASE AYSSSSMSRH 600
601 NTTVAATEHG RNHQQPPSGM VQHHQVYADG NGGSVRVKSE RVATDTATMA FHEQYSNQED 660
661 LMSALLKQEG IAPVDGEFDF DAYSIDNIPV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
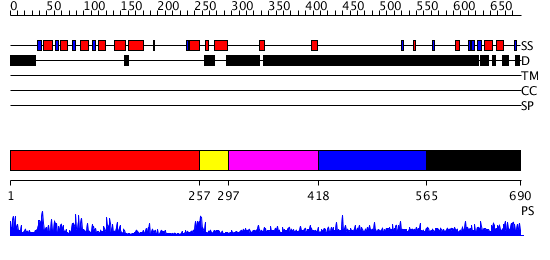
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..256] | 59.69897 | No description for 2oqrA was found. | |
| 2 | View Details | [257..296] | 2.64 | Arr10-B | |
| 3 | View Details | [297..417] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [418..564] | 2.074996 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [565..690] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)