













| General Information: |
|
| Name(s) found: |
JJJ1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 630 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IEA]
|
| Biological Process: |
protein folding
[ISS]
|
| Molecular Function: |
nucleic acid binding
[IEA]
heat shock protein binding [IEA] zinc ion binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASSSRSEKR CHYEVLGISK ESSPDEIRSS YRRLALQRHP DKLMKAAGLS EAEATAQFQE 60
61 LVHAYEVLSD PKERAWYDSH RSQILFADHS SAGGSKSGGS VPDLFAFFSP TVYSGYSDTG 120
121 KGFYKVYYDV FNSVYLNEIK FARTLGLRMD SVREAPIMGN LESPYAQVTA FYNYWLGFCT 180
181 VMDFCWVDEY DVMGGPNRKS RRMMEEENKK SRKKAKREYN DTVRGLAEFV KKRDKRVIDM 240
241 LVKKNAEMEK KKEEERERKK KMEKERLERA MNYEEPEWAK AHEGEDEGAG LSELEEEDDD 300
301 AKRKNEQLYC IVCSKKFKSE KQWKNHEQSK KHKEKVAELR ESFTDYEEEN EEEEIDGPLD 360
361 SPESVEELHE KLQEELNIDN EERDVKKEVV GEADETDDEY FVAEEDMQGS SESEDEDDEM 420
421 TLLKKMVSGQ KNKQKNVVSK EEDEDETEVE IEGDTAEFSE FDNQKSTGRN KEAKEERNKQ 480
481 NAGNDMADDT SKVQIPGEGG NPDENMNATE SASGALADSQ KDEANSMEYD NRKSTGRRRR 540
541 SKKGKDKNNQ GELNEKSSEA DDTQYVNRDM ESQDYKKAPR SKKSTRGMKT KGTTKKNSSN 600
601 ECDRCGEEFE SRTKLHKHLA DSGHATVKSR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
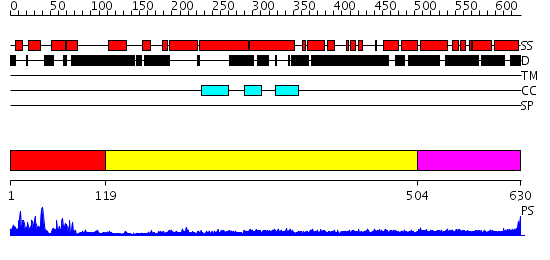
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..118] | 11.154902 | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | |
| 2 | View Details | [119..503] | 23.0 | Heavy meromyosin subfragment | |
| 3 | View Details | [504..630] | 5.39794 | Tropomyosin |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)