













| General Information: |
|
| Name(s) found: |
gi|9758540
[NCBI NR]
gi|15240136 [NCBI NR] |
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 870 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[IDA]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
pyridoxal phosphate binding
[IEA]
catalytic activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHISLWKPIY HCAAALVLDK KSSGSSSSSS RNRDVTQRKL HESKLREALE QASEDGLLVK 60
61 SQDMEEEDES QDQILGRSRS LARLNAQREF LRATSLAAQR AFESEETLPE LEEALTIFLT 120
121 MYPKYQSSEK VDELRNDEYF HLSLPKVCLD YCGFGLFSYL QTVHYWDTCT FSLSEISANL 180
181 SNHAIYGGAE KGSIEHDIKI RIMDYLNIPE NEYGLVFTVS RGSAFKLLAE SYPFHTNKKL 240
241 LTMFDHESQS VSWMGQCAKE KGAKVGSAWF KWPTLRLCSM DLKKEILSKK KRKKDSATGL 300
301 FVFPVQSRVT GSKYSYQWMA LAQQNNWHVL LDAGALGPKD MDSLGLSLFR PDFIITSFYR 360
361 VFGYDPTGFG CLLIKKSVIS CLQSQSGKTS SGIVKITPEY PLYLSDSMDG LEGLTGIQDN 420
421 GIAINGDNKA LGTQLPAFSG AYTSAQVQDV FETDMDHEIG SDRDNTSAVF EEAESISVGE 480
481 LIKSPVFSED ESSDSSLWID LGQSPADSDN AGHLNKQKSP LLVRKNHKRR SSPKPASKAN 540
541 NGSNGGRHVL SFDAAVLSVS HEVGEEVIEE ENSEMNQIDT SRRLRVTEIE EEEEEGGSSK 600
601 LTAHANGSSS GIKDSAIRRE TEGEFRLLGR REKSQYNGGR LLVNEDEHPS KRRVSFRSVD 660
661 HGEASVISLG DEDEEEDGSN GVEWDDDQRE PEIVCRHIDH VNMLGLNKTT SRLRYLINWL 720
721 VTSLLQLRLP RSDSDGEHKN LVQIYGPKIK YERGSSVAFN IRDLKSGMVH PEIVQKLAER 780
781 EGISLGIGYL SHIKIIDNRS EDSSSWKPVD REGRNNGFIR VEVVTASLGF LTNFEDVYRL 840
841 WNFVAKFLSP GFAKQGTLPT VIEEDDSSET |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
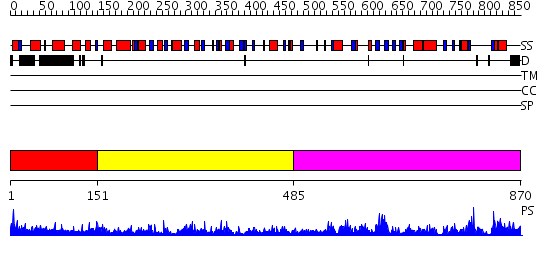
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..150] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [151..484] | 71.39794 | Cysteine desulfurase IscS | |
| 3 | View Details | [485..870] | 66.69897 | NifS-like protein/selenocysteine lyase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 |
|
|||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.62 |
Source: Reynolds et al. (2008)