













| General Information: |
|
| Name(s) found: |
gi|9758415
[NCBI NR]
gi|15240021 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 890 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVRSKAPAKK QQKKGIDFKK IKRKLGRKLP PPKNATNTEI KSKAIILPEQ SVAAEKSGLA 60
61 TSKKGLTLKE LLPQTSHHNA KVRKDALYGI KDLFKNHPEE LQSHKYAIIQ KLRERISDDD 120
121 KLVRDVFYQL FDIDIFPLCK EDNQGLMVSL LMPYIFSAMA HSAIDVRLMA FKFFHLVVEF 180
181 YPPTFSLDAE KILENYKDII QKNHFYVEDK SKLKVALSGL SHCLSVLPCD ESDTKSQKKG 240
241 PSQNETLLTY EQDATKDCVL YSILTKFLRI LTGYAHVYQR LKEVVAVLIN CFQDFIPLIH 300
301 APRGFDAHSF DCIQHILRSI GYAIKFSIRR HKQRQTAWRP ASEEVTLMIL DQDIASMLLK 360
361 KLLGSFPLDP ENNLYGKGDD KYFKLNSVLT EIFLEVSEWS HLPTDLSNRY LEFIENTLLG 420
421 KITRSIRLRK DIHEKTLLAL LPFLPKLILR VDRDWRDNLL EAFTITFSDC KPESKLKLAC 480
481 ISTVRDMIIP NGDILYPNAS DPTVHYYQVA WVNKLPSLLN QLGNEHPVST QVVLQLLLDL 540
541 GRVGCLNASP TCEDDIRNFF SLCQGEGDVP GGPFASLPRE AQELALCFLY YFTTDKFSSP 600
601 MMKAIVSCCL YPQLEPAVLY RIVEILHTAY RAGYIQITDH FSFLITLIAR FKVVPEKLQS 660
661 SIECNERETY CGTFKEVTKL VCSCLSEMGD SSLVLQIVEK VLLEQIVLKP ALDNGCAILR 720
721 MICVLDSTPT TLSERSVTTL SEFLPGYMIN IVNCIPEDKE NSYLYIQTCL YYLVPCYFLF 780
781 NRSSKLTEQV LIRMRSMISE NTKALESVQD RESGRNSLNM IQCIISVILL MHNDVKVRKI 840
841 ISSFKPEIDL ILQNVITLQS SRSTSLTVEG KHMMKIAGER LRIASKSLLA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
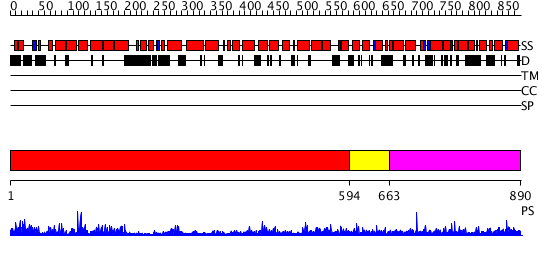
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..593] | 78.69897 | No description for 2qmrA was found. | |
| 2 | View Details | [594..662] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [663..890] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.86 |
Source: Reynolds et al. (2008)