













| General Information: |
|
| Name(s) found: |
ORP1D_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 816 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
steroid metabolic process
[IEA][ISS]
|
| Molecular Function: |
oxysterol binding
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNPLCCIAPV SIDDRTNPVV AKSSNHHLGL EAIPVSKHAS KPSFSTQASW ISQDQLERLS 60
61 SEVVDDVNLD GKDASSSSNK GCFFFGNCVG AGAGVAGIMY KWVNYGKGWR ARWFELEDGV 120
121 LSYYKIHGPD KIVMNPSREK GVRVIGEESV RYIRKASCGS SNRLGASAVA ASRPCKPFGE 180
181 IHLKVSSIRA SKSDDKRLAI FTGTKTLHLR CVSKENRAAW VEAFQVAKDL FPRVASGDIL 240
241 PSEDAVVSTE KLREKLLQEG VGETVVKDCE AIMLSEVSVL QNRLKVLTHK HIILLDTLRQ 300
301 LETEKIELEA TVVDETKEHD SCCGQGRRFS DFYSVMSEVS ASDSEADNES QDGADVESDE 360
361 DDVPYFDTND ILSAEAMRSA SYRSREAEGN GSIYDKDPFF SDRLQIPARI PQYPYVRRRD 420
421 NLPEPKEKEK PVGLWSIIKE NIGKDLSGVC LPVYFNEPLS SLQKCFEDLE YSYLIDRALE 480
481 WGKQGNELMR ILNIAAFAVS GYASTEGRQC KPFNPLLGET YEADYPDKGL RFFSEKVSHH 540
541 PMIVACHCEG QGWNFWGDSN IKGKFWGRSI QLDPVGVLTL KFDDGEIYQW SKVTTSIYNI 600
601 ILGKLYCDHY GTMRIKGGSN YSCRLKFKEQ SVIDRNPRQV HGFVQDNRTG EKVAILIGKW 660
661 DEAMYYVLGD PTTKPKGYDP MTEAVLLWER DKSPTKTRYN LSPFAISLNE ITPGMIDKLP 720
721 PTDSRLRPDQ RHLENGEYES ANAEKLRLEQ LQRQARRLQE KGWKPRWFEK DEEGNYRYLG 780
781 GYWEAREKKD WDRITDIFKK QQQRNSLSSS SSSTFL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
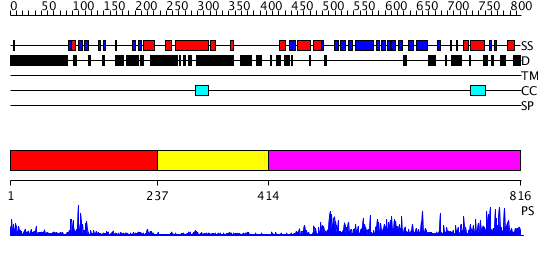
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..236] | 25.154902 | No description for 2r0dA was found. | |
| 2 | View Details | [237..413] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [414..816] | 58.0 | Structure of yeast oxysterol binding protein Osh4 in complex with 7-hydroxycholesterol |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.76 |
Source: Reynolds et al. (2008)