













| General Information: |
|
| Name(s) found: |
MBR1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 704 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: |
zinc ion binding
[IEA][ISS]
protein binding [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNPMQGPRSI GGSSTEVNQV DGESIYCTET SLNTMLNPAD TGFPNNSTPS GRPTYASSSS 60
61 HAAQDHTWWR FGESSSIPGP SDQVNSIGIK TSHQLPQDGT HHFVGYGSEG RQTGLNGMMV 120
121 DGGVHAGSHI RNVPSFLRGS SSNPMPQHVD MSMDMDSDNC NAQTSGVVIR HNSYGSSLGS 180
181 SVQAAGESSS GPASPFGGWG SSCKRKALEG SPSHYFSGET PNRIVQTENS ASHASLSQYG 240
241 ASSSLSLATP SQSSPNVTNH FGRTEQMFGS GGGRAVAASA FHSTRNTDTL SRAGRRLNPR 300
301 QPQESVAFSV SHGGTSVRPT GSLQQNLPLN SPFVDPPDVR SSSITSGSNT GENQTNIVHL 360
361 PALTRNIHQY AWDASFSSRA SNPSGIGMPA ERLGPQWETP RSNQEQPLFA PATDMRQPVH 420
421 DLWNFARGSP GSSVDSLFVP RAGPSSAIHT PQPNPTWIPP QNAPPHNPSR TSELSPWSLF 480
481 PSIESPSASH GGPLPLLPAG PSVSSNEVTM PSSSNSRSHR SRHRRSGLLL ERQNELLHLR 540
541 HIGRSLAADG NGRNQIISEI RQVLHAMRRG ENLRVEDYMV FDPLIYQGMT DMHDRHREMR 600
601 LDVDNMSYEE LLALGERIGD VSTGLSEEVI LKAMKQHKHT SSSPSSVELH QNIEPCCICQ 660
661 EEYVEGDNLG TLKCGHEFHK DCIKQWVMIK NLCPICKTEA LKTP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
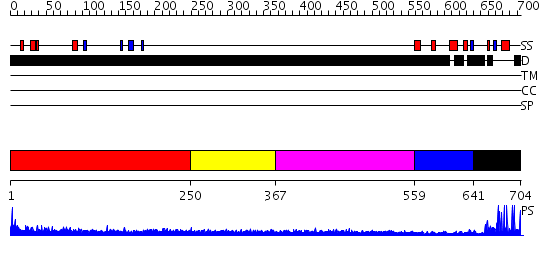
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..249] | 3.234993 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [250..366] | 1.244993 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [367..558] | 2.245994 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [559..640] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [641..704] | 13.0 | No description for 2ep4A was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)