













| General Information: |
|
| Name(s) found: |
gi|9294279
[NCBI NR]
gi|15233054 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 665 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
intracellular signaling pathway
[IEA]
|
| Molecular Function: |
zinc ion binding
[IEA]
protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLRLEKHEH YVDLVKQRDG LECDVCDRSY GDGFYCSECK FTVHRKCAVV FLLKDIFDHP 60
61 SHVGHYLKLL TTGAPDHTDP KCHLCGKNTK RILYHCSICK LNLDIDCIFD YFSDQQAHLN 120
121 FSWHHHPLHF ISFRLHLQCD VCYMFRRKGY FCFQCKLVVH KECVSKIESA VITHPFHARH 180
181 PLKLLTDGAP DYTDPKCHIC GVETANLLYH CDMCMFNLDI GCAIRNPERV SLSKLKVHEH 240
241 TLTLMPKLMF FVCDACGMKG DHAPYVCVQC DFMFFHQKCA YLPRVINVNH HEHRVSYKYP 300
301 LGPGDWICGV CWEEINWSYG AYSCSSCPNY AIHSRCATRK DVWDGKELDG VPELIEDIKP 360
361 FKMNDDNTIT HFAHGHNLSL NKDTEEGSFC EACVLPIDSY TSYKCSKSDC CFILHETCAN 420
421 FPKKKRHFLS PEPLTLRPQN QRKKESICRA CDQVFCQGFV YSTYRKRNFD LLCSSITMPF 480
481 KHRSHDHHLL YLKLEVDHTK TCQNCGIDEK EVVLGCLKCN YFLDFRCATL PYTVRLPRYD 540
541 DHPLTLCYGE KASGKYWCDI CERETNSKTW FYTCKGCGVT LHVFCVLGDI RYAKPGGSIN 600
601 NNVELVPNNR SSRPLCHNCH CHCPGPFILK DFVEECFCSY YCFSRCKSWM SIVKKEKGIC 660
661 PPWAL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
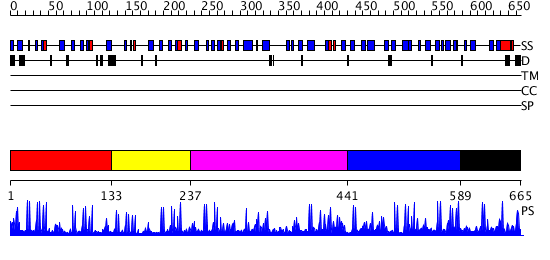
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..132] | 4.30103 | Crystal Structure of the Human Beta2-Chimaerin | |
| 2 | View Details | [133..236] | 2.45 | Solution Structure of DC1 Domain of PDI-like Hypothetical Protein from Arabidopsis thaliana | |
| 3 | View Details | [237..440] | 2.152987 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [441..588] | 3.09691 | No description for 2ysmA was found. | |
| 5 | View Details | [589..665] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)