













| General Information: |
|
| Name(s) found: |
PARG1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 548 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
defense response to fungus
[IMP]
|
| Molecular Function: |
poly(ADP-ribose) glycohydrolase activity
[IMP][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MENREDLNSI LPYLPLVIRS SSLYWPPRVV EALKAMSEGP SHSQVDSGEV LRQAIFDMRR 60
61 SLSFSTLEPS ASNGYAFLFD ELIDEKESKR WFDEIIPALA SLLLQFPSLL EVHFQNADNI 120
121 VSGIKTGLRL LNSQQAGIVF LSQELIGALL ACSFFCLFPD DNRGAKHLPV INFDHLFASL 180
181 YISYSQSQES KIRCIMHYFE RFCSCVPIGI VSFERKITAA PDADFWSKSD VSLCAFKVHS 240
241 FGLIEDQPDN ALEVDFANKY LGGGSLSRGC VQEEIRFMIN PELIAGMLFL PRMDDNEAIE 300
301 IVGAERFSCY TGYASSFRFA GEYIDKKAMD PFKRRRTRIV AIDALCTPKM RHFKDICLLR 360
361 EINKALCGFL NCSKAWEHQN IFMDEGDNEI QLVRNGRDSG LLRTETTASH RTPLNDVEMN 420
421 REKPANNLIR DFYVEGVDNE DHEDDGVATG NWGCGVFGGD PELKATIQWL AASQTRRPFI 480
481 SYYTFGVEAL RNLDQVTKWI LSHKWTVGDL WNMMLEYSAQ RLYKQTSVGF FSWLLPSLAT 540
541 TNKAIQPP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
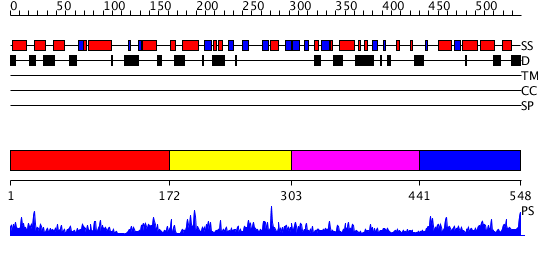
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..171] | 1.053969 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [172..302] | 1.061956 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [303..440] | 1.066967 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [441..548] | 9.042992 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.79 |
Source: Reynolds et al. (2008)