













| General Information: |
|
| Name(s) found: |
Y5880_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 614 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[IDA]
|
| Biological Process: |
response to light stimulus
[IEA][ISS]
|
| Molecular Function: |
protein binding
[IEA]
signal transducer activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDKHHHQHLL LQHHQHLHHH QKLSLAKSSR QSCSEWIFRD VPSDITIEVN GGNFALHKFP 60
61 LVSRSGRIRR IVAEHRDSDI SKVELLNLPG GAETFELAAK FCYGINFEIT SSNVAQLFCV 120
121 SDYLEMTEEY SKDNLASRTE EYLESIVCKN LEMCVQVLKQ SEILLPLADE LNIIGRCIDA 180
181 IASKACAEQI ASSFSRLEYS SSGRLHMSRQ VKSSGDGGDW WIEDLSVLRI DLYQRVMNAM 240
241 KCRGVRPESI GASLVSYAER ELTKRSEHEQ TIVETIVTLL PVENLVVPIS FLFGLLRRAV 300
301 ILDTSVSCRL DLERRLGSQL DMATLDDLLI PSFRHAGDTL FDIDTVHRIL VNFSQQGGDD 360
361 SEDEESVFEC DSPHSPSQTA MFKVAKLVDS YLAEIAPDAN LDLSKFLLIA EALPPHARTL 420
421 HDGLYRAIDL YLKAHQGLSD SDKKKLSKLI DFQKLSQEAG AHAAQNERLP LQSIVQVLYF 480
481 EQLKLRSSLC SSYSDEEPKP KQQQQQSWRI NSGALSATMS PKDNYASLRR ENRELKLELA 540
541 RLRMRLNDLE KEHICMKRDM QRSHSRKFMS SFSKKMGKLS FFGHSSSRGS SSPSKQSFRT 600
601 DSKVLMERTC ASTD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
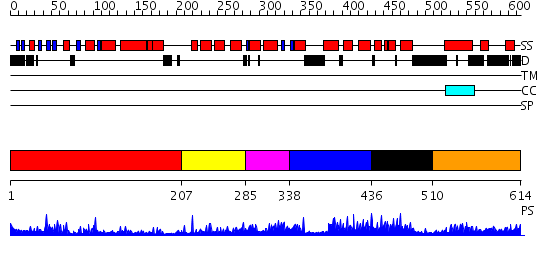
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..206] | 21.0 | No description for 2nn2A was found. | |
| 2 | View Details | [207..284] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [285..337] | 4.045757 | No description for 2dyhA was found. | |
| 4 | View Details | [338..435] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [436..509] | 3.077994 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [510..614] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)