













| General Information: |
|
| Name(s) found: |
gi|67633364
[NCBI NR]
gi|25402640 [NCBI NR] gi|1931643 [NCBI NR] gi|15220265 [NCBI NR] |
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 438 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
protein folding
[IEA][ISS]
|
| Molecular Function: |
heat shock protein binding
[IEA]
unfolded protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGTPRSPATD FFGISKTACK AYKSLVTKLH PLSSHRKSES HSDADSAIHR RYLEEKFAEE 60
61 DDLIAARRGL RLQSMDDSSV FKRRSSLLSN SSSRRSHTPQ ARPTYLSSSA SSNRRSAFSR 120
121 STSRRDEGSS HSGVRSHGLK SRPISNNTSP MTSPFTSPRG KDQTLGDLFG SVKKLSSPTS 180
181 NSPINGVKQS SPSSISKSAS KRDKDERGST SSATSTSLPY SKSKSTRDTA GSIAKSISRR 240
241 STTPIVFSQS TPPKKPPAVE KKLECTLEEL CHGGVKNIKI KRDIITDEGL IMQQEEMLRV 300
301 NIQPGWKKGT KITFEGVGNE KPGYLPEDIT FVVEEKRHPL FKRRGDDLEI AVEIPLLKAL 360
361 TGCKLSVPLL SGESMSITVG DVIFHGFEKA IKGQGMPNAK EEGKRGDLRI TFLVNFPEKL 420
421 SEEQRSMAYE VLKDCSWA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
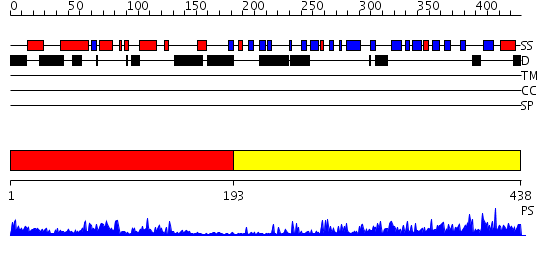
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..192] | 19.522879 | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | |
| 2 | View Details | [193..438] | 43.522879 | The crystal structure of Hsp40 Ydj1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)