













| General Information: |
|
| Name(s) found: |
gi|3608134
[NCBI NR]
gi|25408387 [NCBI NR] gi|15227436 [NCBI NR] |
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 575 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
protein folding
[ISS]
|
| Molecular Function: |
heat shock protein binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASGSDSEVE KESIHHKALA ESSFNCGDLM SALTHARKAL SLSPNTEGLS AMVTAFEIIS 60
61 SAATVAGGFP EWYKVLKVEP FSHINTIKQQ YRKLALVLHP DKNPYVGCEE GFKLLNEAFR 120
121 VFSDKGEMVS GGSGGDETST FSAVCSGCRS VHKFDRKYLG QNLMCPTCKN SFEAKEVEKE 180
181 EEGRENGACT SKIITYSRRK RPVDSDGESL IKEVETGEMS QEAAEAINVF EGSDEEDEGM 240
241 MTLAEMQAVI KRNKSKVNSK ITEKDSSGEE NMGRETQKRS LADASMTETL REMSTNAVNN 300
301 KQEVLKNRKN IKKKKMANHK DLTEIVDLEY VPRVDRKRDR GKLSQEMYME DEDFELYDFD 360
361 KDRMPRSFKK GQIWAIYDGG DDKMPRSYCL VSEVVSLNPF KVWISWLDFE SEKLISWMKI 420
421 SSSHMPCGRF RVSEKALIEQ VKPFSHLVNC ERAAREIYQI YPKKGSVWAV YSETNPGLQR 480
481 RKTRRYEIVV CLTMYTDAYG LSVAYLEKVN DYSNLFKRRD YGYNAVRWVE KEDVAALLSH 540
541 QIPAKKLPED ESGADLKESW VLDLASVPPD LVSAT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
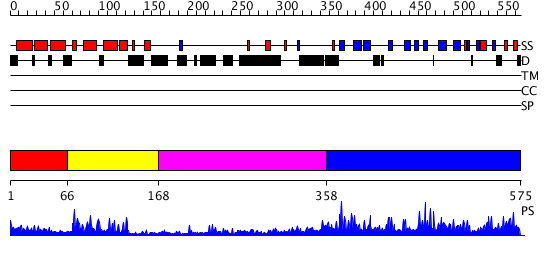
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..65] | 1.270996 | View MSA. Confident ab initio structure predictions are available. | |
| 2 | View Details | [66..167] | 22.69897 | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | |
| 3 | View Details | [168..357] | 21.0 | The crystal structure of Hsp40 Ydj1 | |
| 4 | View Details | [358..575] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)