













| General Information: |
|
| Name(s) found: |
PNSL3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 220 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
chloroplast photosystem II [ISS] oxygen evolving complex [TAS] chloroplast thylakoid lumen [IDA] chloroplast thylakoid membrane [IDA] |
| Biological Process: |
photosynthesis, light reaction
[TAS]
|
| Molecular Function: |
calcium ion binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAHFIDLNSL TNTLPSLPKL PESRKTGKSS GFACRRTEEF QEPDSVQITR RMTLGFAVSI 60
61 GLTGILGENN VSLAQDNGFW IDGPLPIPPI YNNIVNEKTG TRTFIKKGVY VADIGTKGRM 120
121 YRVKKNAFDL LAMEDLIGPD TLNYVKKYLR LKSTFLFYDF DNLISAAASE DKQPLTDLAN 180
181 RLFDNFEKLE DAAKTKNLAE TESCYKDTKF LLQEVMTRMA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
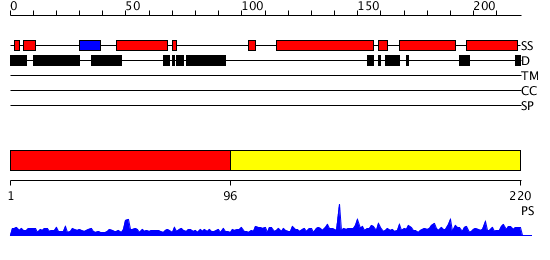
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..95] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [96..220] | 42.69897 | Crystal structure of PsbQ polypeptide of photosystem II from higher plants |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)