













| General Information: |
|
| Name(s) found: |
gi|9759539
[NCBI NR]
gi|15242956 [NCBI NR] gi|110738487 [NCBI NR] |
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 748 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
gene silencing
[IMP]
DNA demethylation [IMP] |
| Molecular Function: |
nucleic acid binding
[ISS]
RNA binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEEKSSGGGV RLHVGGLGES VGRDDLLKIF SPMGTVDAVE FVRTKGRSFA YIDFSPSSTN 60
61 SLTKLFSTYN GCVWKGGRLR LEKAKEHYLA RLKREWEAAS STSDNTIKAP SDSPPATHLN 120
121 IFFPRLRKVK PMPLSGTGKH KYSFQRVPVS SSLPRSFCDC EEHSNSSLTP REIHLHDLEA 180
181 VNVGRQEAEV NVMNSVMNKL FEKNNVDPEE DNEIEADQDN LIINVASSGN DMDSALDMLS 240
241 RKRKSILNKK TPSEEGYSEG RKGNLTHPSK NRQTISLEET GRQESSQAIR GKKKPSEVVP 300
301 DKSSDEPSRT KDLEQSIDNI SWSQKSSWKS LMANGNSNDF SVSSFLPGVG SSKAVQPAPR 360
361 NTDLAGLPSR ENLKKKTKRK RVTSTIMAED LPVSDDIKRD DSDTMADDIE RDDSDAVEYY 420
421 TACESMADDT ASDSVAERDD SDAVEDDTAI DSMADDPASD SVAESDDGDA VENDTAIDSM 480
481 ADDTVSNSMA ESDDGDNVED DTAIDSMCDD TANDDVGSDD SGSLADTVSD TSVEAVPLEF 540
541 VANTEGDSVD GKSNVEKHEN VAEDLNAEKE SLVVKENVVD EEEAGKGPLK ASNKSTGGSS 600
601 WLQKASWTQL VSDKNTSSFS ITQLFPDLTS DKGEAAGVIN NVGNQFSNSN QTASAMKQTD 660
661 YASSSGGFVA AGVPVDSTPV RSLDENRQRL NGKNVSEGAK LGAKKKIIKR KVGSGDTCTF 720
721 MRSSTSLKEW AKAKKALSEP RRKKNSEE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
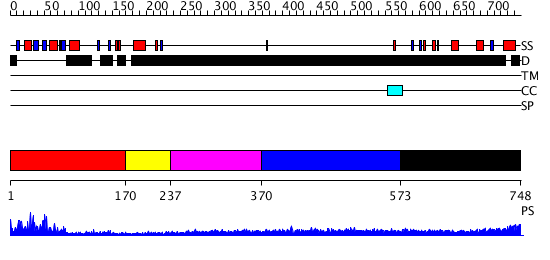
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..169] | 28.09691 | Poly(A)-binding protein | |
| 2 | View Details | [170..236] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [237..369] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [370..572] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [573..748] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)