













| General Information: |
|
| Name(s) found: |
gi|51971657
[NCBI NR]
gi|42565086 [NCBI NR] gi|11994389 [NCBI NR] |
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 458 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IEA]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
hydrolase activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLCVFRRKLG CLFSRLRWVI KKRVRARVIV RRFRKARWRA RRKESPESEV SSIHLSSNSG 60
61 RHIRVATFNV AMFSLAPVVQ TMEETAFLGH LDSSNITCPS PKGILKQSPL HSSAVRKPKV 120
121 CINLPDNEIS LAQSYSFLSM VENDNDGKEN RGSLSMRSPV CLPSCWWDQE SFNGYSSRRS 180
181 IAELLRELDA DILALQDVKA EEETLMKPLS DLASALGMKY VFAESWAPEY GNAILSKWPI 240
241 KKWRVQRIAD VDDFRNVLKV TVEIPWAGDV NVYCTQLDHL DENWRMKQID AITRGDESPH 300
301 ILLGGLNSLD GSDYSIARWN HIVKYYEDSG KPTPRVEVMR FLKGKGYLDS KEFAGECEPV 360
361 VIIAKGQNVQ GTCKYGTRVD YILASPESPY EFVPGSYSVV SSKGTSDHHI VKVDLVITKE 420
421 RSRGNFKHSR KKAKQKIFQI KANLMSKDTW KLGNLMSS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
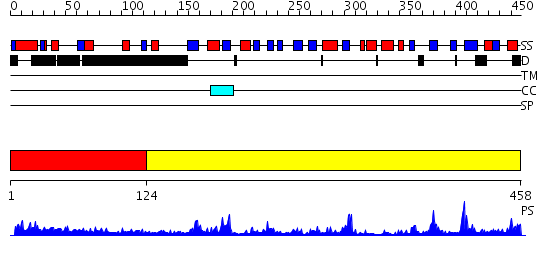
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..123] | 1.213994 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [124..458] | 56.39794 | THE CRYSTAL STRUCTURE OF THE HUMAN DNA REPAIR ENDONUCLEASE HAP1 SUGGESTS THE RECOGNITION OF EXTRA-HELICAL DEOXYRIBOSE AT DNA ABASIC SITES |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||
| 1 | No functions predicted. | ||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.82 |
Source: Reynolds et al. (2008)