













| General Information: |
|
| Name(s) found: |
SWI3C_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 807 amino acids |
Gene Ontology: |
|
| Cellular Component: |
SWI/SNF complex
[ISS]
chromatin remodeling complex [ISS] |
| Biological Process: |
chromatin remodeling
[ISS]
|
| Molecular Function: |
DNA binding
[IEA][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPASEDRRGK WKRKKRGGLS AARKPKQEEE DMEEEDEENN NNNNEEMDDV ENADELQQNG 60
61 GATPDPGLGI GEVVEDSGSR ISDFPAVVKR VVIRPHASVM AVVAAERAGL IGETRGQGSL 120
121 PALENISFGQ LQALSTVPAD SLDLERSDGS SSAYVISPPP IMDGEGVVKR FGDLVHVLPM 180
181 HSDWFAPNTV DRLERQVVPQ FFSGKSPNHT PESYMEFRNA IVSKYVENPE KTLTISDCQG 240
241 LVDGVDIEDF ARVFRFLDHW GIINYCATAQ SHPGPLRDVS DVREDTNGEV NVPSAALTSI 300
301 DSLIKFDKPN CRHKGGEVYS SLPSLDGDSP DLDIRIREHL CDSHCNHCSR PLPTVYFQSQ 360
361 KKGDILLCCD CFHHGRFVVG HSCLDFVRVD PMKFYGDQDG DNWTDQETLL LLEAVELYNE 420
421 NWVQIADHVG SKSKAQCILH FLRLPVEDGL LDNVEVSGVT NTENPTNGYD HKGTDSNGDL 480
481 PGYSEQGSDT EIKLPFVKSP NPVMALVAFL ASAVGPRVAA SCAHESLSVL SEDDRMKSEG 540
541 MQGKEASLLD GENQQQDGAH KTSSQNGAEA QTPLPQDKVM AAFRAGLSAA ATKAKLFADH 600
601 EEREIQRLSA NIVNHQLKRM ELKLKQFAEI ETLLMKECEQ VEKTRQRFSA ERARMLSARF 660
661 GSPGGISPQT NNLQGMSLST GGNNINSLMH QQHQQQQASA TSQPSIIPGF SNNPQVQAQM 720
721 HFMARQQQQQ QQQQQQQQQA FSFGPRLPLN AIQTNAGSTA SPNVMFGNNQ LNNPAAAGAA 780
781 SINQPSFSHP MVRSSTGSGS GSGLGLN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
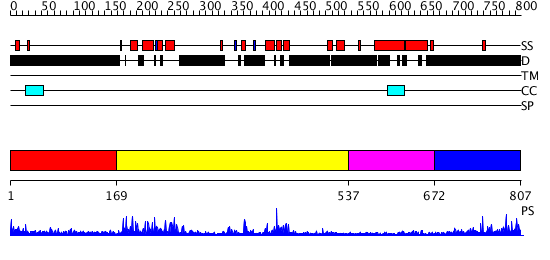
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..168] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [169..536] | 51.221849 | No description for 2dw4A was found. | |
| 3 | View Details | [537..671] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [672..807] | 5.206996 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)