













| General Information: |
|
| Name(s) found: |
P4KG3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 574 amino acids |
Gene Ontology: |
|
| Cellular Component: |
peroxisome
[IDA]
|
| Biological Process: |
protein modification process
[IEA]
|
| Molecular Function: |
inositol or phosphatidylinositol kinase activity
[ISS]
phosphotransferase activity, alcohol group as acceptor [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSVASVALSP ALEELVNFPG IIGRFGFNLD DPILVFLTIA GSVIPKRVME SDSIASVKLR 60
61 IQSIKGFFVK KQKLLYDGRE VSRNDSQIRD YGLADGKLLH LVIRLSDLQA ISVRTVDGKE 120
121 FELVVERSRN VGYVKQQIAS KEKELGIPRD HELTLDGEEL DDQRLITDLC QNGDNVIHLL 180
181 ISKSAKVRAK PVGKDFEVFI EDVNHKHNVD GRRGKNISSE AKPKEFFVEP FIVNPEIKLP 240
241 ILLKELISST LEGLEKGNGP IRSSDGSGGA YFMQDPSGHK YVSVFKPIDE EPMAVNNPHG 300
301 QPVSVDGEGL KKGTQVGEGA IREVAAYILD YPMTGPRTFP HDQTGFAGVP PTTMVKCLHK 360
361 DFNHPNGYSF SPENTKIGSL QMFVSNVGSC EDMGYRVFPV DQVHKISVLD IRLANADRHA 420
421 GNILVSRDGK DGQMVLTPID HGYCFPNKFE DCTFEWLYWP QAKEPYSSET LEYIKSLDPE 480
481 KDIELLRFHG WEIPPSCTRV LRISTMLLKK GSAKGLTPFT IGSIMCRETL KEESVIEQII 540
541 HDAEAIVPTE TTEDEFISTV SAIMDNRLDQ YAWN |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
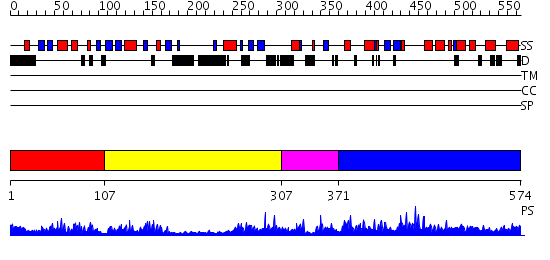
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..106] | 22.09691 | Crystal Structure of ISG15, the Interferon-Induced Ubiquitin Cross Reactive Protein | |
| 2 | View Details | [107..306] | 29.09691 | Structure of the DNA repair protein hHR23a | |
| 3 | View Details | [307..370] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [371..574] | 6.057977 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 |
|
|||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)