













| General Information: |
|
| Name(s) found: |
FRS5_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 788 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
response to red or far red light
[ISS]
|
| Molecular Function: |
zinc ion binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMDNEVLDFD IGVGVSSGGD VDDDAIDIEH HALDDDDMLD SPIMPCGNGL VGNSGNYFPN 60
61 QEEEACDLLD LEPYDGLEFE SEEAAKAFYN SYARRIGFST RVSSSRRSRR DGAIIQRQFV 120
121 CAKEGFRNMN EKRTKDREIK RPRTITRVGC KASLSVKMQD SGKWLVSGFV KDHNHELVPP 180
181 DQVHCLRSHR QISGPAKTLI DTLQAAGMGP RRIMSALIKE YGGISKVGFT EVDCRNYMRN 240
241 NRQKSIEGEI QLLLDYLRQM NADNPNFFYS VQGSEDQSVG NVFWADPKAI MDFTHFGDTV 300
301 TFDTTYRSNR YRLPFAPFTG VNHHGQPILF GCAFIINETE ASFVWLFNTW LAAMSAHPPV 360
361 SITTDHDAVI RAAIMHVFPG ARHRFCKWHI LKKCQEKLSH VFLKHPSFES DFHKCVNLTE 420
421 SVEDFERCWF SLLDKYELRD HEWLQAIYSD RRQWVPVYLR DTFFADMSLT HRSDSINSYF 480
481 DGYINASTNL SQFFKLYEKA LESRLEKEVK ADYDTMNSPP VLKTPSPMEK QASELYTRKL 540
541 FMRFQEELVG TLTFMASKAD DDGDLVTYQV AKYGEAHKAH FVKFNVLEMR ANCSCQMFEF 600
601 SGIICRHILA VFRVTNLLTL PPYYILKRWT RNAKSSVIFD DYNLHAYANY LESHTVRYNT 660
661 LRHKASNFVQ EAGKSLYTCD VAVVALQEAA KTVSLAMNKE VRRTMANRHF KASSVTGGKH 720
721 QQEVLAQPEP EDEMDKKINQ LRNELELANR KCEAYRTNLL SVLKEMEDQK LQVSIKVQNI 780
781 KISLKDNL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
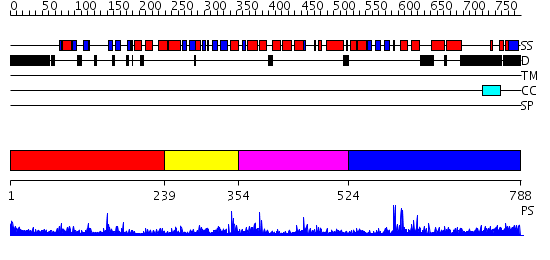
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..238] | 5.299992 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [239..353] | 2.395996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [354..523] | 9.402994 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [524..788] | 6.231988 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)