













| General Information: |
|
| Name(s) found: |
gi|9294595
[NCBI NR]
gi|51970142 [NCBI NR] gi|51969228 [NCBI NR] gi|46518487 [NCBI NR] gi|15233233 [NCBI NR] |
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 755 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
intracellular signaling pathway
[ISS]
signal transduction [ISS] |
| Molecular Function: |
phosphoinositide binding
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNLYAHDLSL LDFNYNVSGP FGEPLSHRFL SPGPFFQGEE DDYRRNTSYS HDGANGREPD 60
61 TDSRASPPHR HDGRSPLPLG MDWSSPPRHL EGRDTVWPHD HRTGWSYCVT VPSWVDLPKS 120
121 SVSDPAVFYR VQVAIQSPEG ITSARLVLRR FNDFLELYSS IKKEFVKKSL PQAPPKKILR 180
181 MRNQTLLEER RCSLEDWMNR LLSDIDISRS ALIATFLELE AAVRSYFNDE YQETEDTSGD 240
241 IPSLLPTTIS DVPGSSSVTI DHDNDSADET SNASTMKHDE ANLKNLFSRN STAVDNVTDW 300
301 HELITEYGLL DQSSFQEKVE RLSSTNGDAA TGTVTRGGIS SGVGIQRLDG SDRKFQELTI 360
361 ESIKKTHVSD FEASTAVEPD LVNQGAMDIH GEAHGNMYGA VGGDTETQKD LAIVFQSEER 420
421 HKLKRVIDTL KQRLETAKAD TEDLISRLNQ ELAVRQFLST KVRDLEVELE TTRESCKQGM 480
481 EKTVLDEKER FTQIQWDMEE LRKQCMEMES FLNSIKDEKT HIETANESLV QENQMLLQQI 540
541 NDIRENFENF HKEHEELEVK AKAELKVLVK EVKSLRTTQS DLRQELSGIM KEKLEMERIV 600
601 QREKDREETA KNADKKLLHE CDVLQNRLQE CNVKFDIEEE GKLIMDSSSL SEAIELLATS 660
661 DNRIGLLIAE TQLLSEEVEK LKLTSGGHRG TDDLVRKMLT EVLIDNARLR KQVNSVLRCS 720
721 LSGHGISVRE AGTEVDDEEG SIDLARTVMS KILEK |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
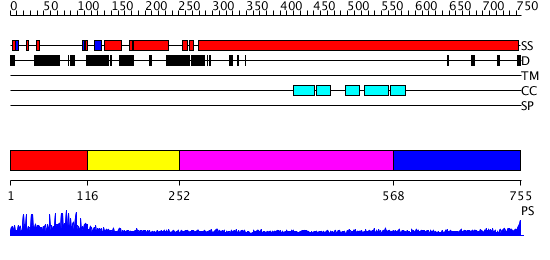
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..115] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [116..251] | 6.045757 | No description for 2i1jA was found. | |
| 3 | View Details | [252..567] | 17.0 | Heavy meromyosin subfragment | |
| 4 | View Details | [568..755] | 6.69897 | No description for 2dfsA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)