













| General Information: |
|
| Name(s) found: |
KCS9_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 512 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[IEA]
|
| Biological Process: |
response to cold
[IEP]
very long-chain fatty acid metabolic process [IDA] cuticle development [IDA] response to light stimulus [IEP] |
| Molecular Function: |
transferase activity, transferring acyl groups other than amino-acyl groups
[IEA]
catalytic activity [IEA] acyltransferase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEAANEPVNG GSVQIRTENN ERRKLPNFLQ SVNMKYVKLG YHYLITHLFK LCLVPLMAVL 60
61 VTEISRLTTD DLYQIWLHLQ YNLVAFIFLS ALAIFGSTVY IMSRPRSVYL VDYSCYLPPE 120
121 SLQVKYQKFM DHSKLIEDFN ESSLEFQRKI LERSGLGEET YLPEALHCIP PRPTMMAARE 180
181 ESEQVMFGAL DKLFENTKIN PRDIGVLVVN CSLFNPTPSL SAMIVNKYKL RGNVKSFNLG 240
241 GMGCSAGVIS IDLAKDMLQV HRNTYAVVVS TENITQNWYF GNKKAMLIPN CLFRVGGSAI 300
301 LLSNKGKDRR RSKYKLVHTV RTHKGAVEKA FNCVYQEQDD NGKTGVSLSK DLMAIAGEAL 360
361 KANITTLGPL VLPISEQILF FMTLVTKKLF NSKLKPYIPD FKLAFDHFCI HAGGRAVIDE 420
421 LEKNLQLSQT HVEASRMTLH RFGNTSSSSI WYELAYIEAK GRMKKGNRVW QIAFGSGFKC 480
481 NSAVWVALNN VKPSVSSPWE HCIDRYPVKL DF |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
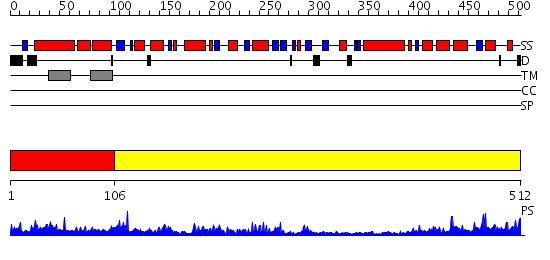
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..105] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [106..512] | 87.0 | Chalcone synthase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||
| 2 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|