













| General Information: |
|
| Name(s) found: |
ATE2_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 605 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
regulation of lipid catabolic process
[IGI]
response to abscisic acid stimulus [IGI] regulation of seed germination [IGI] protein arginylation [ISS] |
| Molecular Function: |
arginyltransferase activity
[IEA][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSKKAKTKN EASSSRGGIG GESIVADCGR NKSTCGYCKS STRFSISHGL WTERLTVNDY 60
61 QALLDSGWRR SGCYLYKPEM EKTCCPSYTI RLKASDFVPS KEQQRVRRRI ERFLDGELDA 120
121 KPSEQTEDQD VSFSREVSVS VKKSLGAAKR EKNNELEPIM KDLSEQIDNA VQKCIQSGEF 180
181 PSNVQIPKAS VKKVFSAKRK KLAEGSEDLL YTSNIAFPIV AAMKHTQTLE KGKNVEENRL 240
241 SPEAVSEKLL SAMNKVGEFT GFSVKVSKGH INFLSATQVT SSDRNEGEES LCATTIKSSS 300
301 NKLHARKRKL EMHLKRSSFE PEEYELYKRY QMKVHNDKPE SISETSYKRF LVDTPLTEVP 360
361 SSGYDDEEKI PLCGFGSFHQ QYRVDDRLIA VGVIDILPKC LSSKYLFWDP DFASLSLGNY 420
421 SALQEIDWVK QNQAHCSTLE YYYLGYYIHS CNKMRYKAAY RPSELLCPLR YQWVPFEVAK 480
481 PLLDKKPYSV LSNISKVSSS SSSPQASETL LESTSEHEDM EQGDTNDDDD EMYNSDEDSD 540
541 SDSSSSRNRS DITNILISLN GPRLRYKDIP RFKNPVVQKQ LESMLVSYRK VVGAELSEIM 600
601 VYELR |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
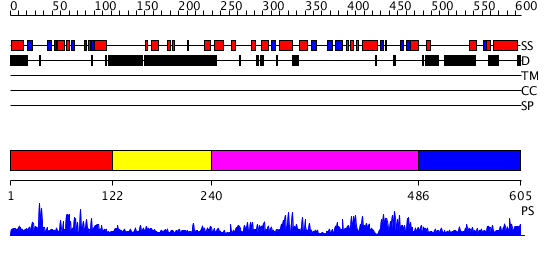
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..121] | 49.142668 | No description for PF04376.4 was found. No confident structure predictions are available. | |
| 2 | View Details | [122..239] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [240..485] | 107.229148 | No description for PF04377.6 was found. No confident structure predictions are available. | |
| 4 | View Details | [486..605] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)