













| General Information: |
|
| Name(s) found: |
PUM2_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 972 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
RNA binding
[IDA][ISS]
mRNA binding [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIPELGRRPM HRGNEDSSFG DDYEKEIGVL LGEQQRRQVE ADELERELNL YRSGSAPPTV 60
61 DGSVSAAGGL FSGGGGAPFL EFGGGNKGNG FGGDDEEFRK DPAYLSYYYA NMKLNPRLPP 120
121 PLMSREDLRV AQRLKGSSNV LGGVGDRRKV NDSQSLFSMP PGFDQRKQHE FEVEKTSASS 180
181 SEWDANGLIG LPGLGIGGKQ KSFADIFQAD MGHGHPVTKQ PSRPASRNTF DENVDSKNNL 240
241 SPSASQGIGA PSPYSYAAVL GSSLSRNGTP DPQAIARVPS PCLTPIGSGR VSSNDKRNTS 300
301 NQSPFNGGLN ESSDLVNALS GMNLSGSGGL DERGQAEQDV EKVRNYMFGL QGGHNEVNQH 360
361 GFPNKSDQAQ KATGLLRNSQ LRGAQGSTYN DGGGVATQYQ HLDSPNYCLN NYGLNPAVAS 420
421 MMANQLGTNN YSPVYENASA ASAMGFSGMD SRLHGGGYVS SGQNLSESRN LGRFSNRMMG 480
481 GGTGLQSHMA DPMYHQYADS LDLLNDPSMD VNFMGNSYMN MLELQRAYLG AQKSQYGVPY 540
541 KSGSPNSHTD YGSPTFGSYP GSPLAHHLLP NSLVSPCSPM RRGEVNMRYP SATRNYAGGV 600
601 MGSWHMDASL DEGFGSSMLE EFKSNKTRGF ELAEIAGHVV EFSSDQYGSR FIQQKLETAT 660
661 SDEKNMVYEE IMPHALALMT DVFGNYVIQK FFEHGLPPQR RELADKLFDN VLPLSLQMYG 720
721 CRVIQKAIEV VDLDQKIKMV KELDGHVMRC VRDQNGNHVV QKCIECVPEE NIEFIISTFF 780
781 GNVVTLSTHP YGCRVIQRVL EHCHDPDTQS KVMDEIMSTI SMLAQDQYGN YVIQHVLEHG 840
841 KPDERTVIIK ELAGKIVQMS QQKFASNVVE KCLTFGGPEE REFLVNEMLG TTDENEPLQA 900
901 MMKDQFANYV VQKVLETCDD QQRELILGRI KVHLNALKKY TYGKHIVARV EKLVAAGERR 960
961 MALQSLTQPQ MA |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
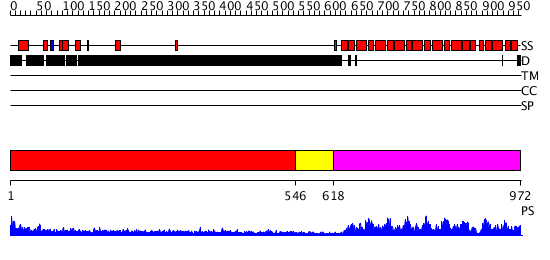
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..545] | 3.30103 | No description for 1y0fB was found. | |
| 2 | View Details | [546..617] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [618..972] | 147.0 | Pumilio 1 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)