













| General Information: |
|
| Name(s) found: |
TPS9_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 867 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
metabolic process
[IEA]
trehalose biosynthetic process [IEA][ISS] |
| Molecular Function: |
transferase activity, transferring glycosyl groups
[ISS]
trehalose-phosphatase activity [IMP][ISS] [NOT]alpha,alpha-trehalose-phosphate synthase (UDP-forming) activity [IMP] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVSRSCANFL DLASWDLLDF PQTQRALPRV MTVPGIISEL DGGYSDGSSD VNSSNSSRER 60
61 KIIVANMLPL QAKRDTETGQ WCFSWDEDSL LLQLRDGFSS DTEFVYIGSL NADIGISEQE 120
121 EVSHKLLLDF NCVPTFLPKE MQEKFYLGFC KHHLWPLFHY MLPMFPDHGD RFDRRLWQAY 180
181 VSANKIFSDR VMEVINPEED YVWIHDYHLM VLPTFLRKRF NRIKLGFFLH SPFPSSEIYR 240
241 TLPVRDDLLR GLLNCDLIGF HTFDYARHFL SCCSRMLGLD YESKRGHIGL DYFGRTVFIK 300
301 ILPVGIHMGR LESVLNLPST AAKMKEIQEQ FKGKKLILGV DDMDIFKGIS LKLIAMERLF 360
361 ETYWHMRGKL VLIQIVNPAR ATGKDVEEAK KETYSTAKRI NERYGSAGYQ PVILIDRLVP 420
421 RYEKTAYYAM ADCCLVNAVR DGMNLVPYKY IICRQGTPGM DKAMGISHDS ARTSMLVVSE 480
481 FIGCSPSLSG AIRVNPWDVD AVAEAVNLAL TMGETEKRLR HEKHYHYVST HDVGYWAKSF 540
541 MQDLERACRE HYNKRCWGIG FGLSFRVLSL SPSFRKLSID HIVSTYRNTQ RRAIFLDYDG 600
601 TLVPESSIIK TPNAEVLSVL KSLCGDPKNT VFVVSGRGWE SLSDWLSPCE NLGIAAEHGY 660
661 FIRWSSKKEW ETCYSSAEAE WKTMVEPVMR SYMDATDGST IEYKESALVW HHQDADPDFG 720
721 ACQAKELLDH LESVLANEPV VVKRGQHIVE VKPQGVSKGL AVEKVIHQMV EDGNPPDMVM 780
781 CIGDDRSDED MFESILSTVT NPDLPMPPEI FACTVGRKPS KAKYFLDDVS DVLKLLGGLA 840
841 AATSSSKPEY QQQSSSLHTQ VAFESII |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
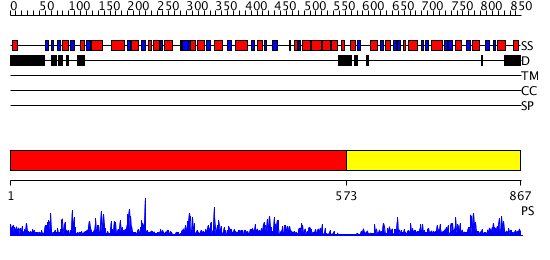
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..572] | 82.69897 | Trehalose-6-phosphate from E. coli bound with UDP-2-fluoro glucose. | |
| 2 | View Details | [573..867] | 26.69897 | Crystal structure of NYSGRC target T1436: A Hypothetical protein yidA. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)