













| General Information: |
|
| Name(s) found: |
SAE2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 700 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA]
|
| Biological Process: |
protein sumoylation
[IDA][ISS]
embryonic development ending in seed dormancy [IMP] |
| Molecular Function: |
SUMO activating enzyme activity
[IDA][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATQQQQSAI KGAKVLMVGA GGIGCELLKT LALSGFEDIH IIDMDTIEVS NLNRQFLFRR 60
61 SHVGQSKAKV ARDAVLRFRP NINIRSYHAN VKNPEFDVDF FKQFDVVLNG LDNLDARRHV 120
121 NRLCLAADVP LVESGTTGFL GQVTVHIKGK TECYECQTKP APKTYPVCTI TSTPTKFVHC 180
181 IVWAKDLLFA KLFGDKNQDN DLNVRSNNSA SSSKETEDVF ERSEDEDIEQ YGRKIYDHVF 240
241 GSNIEAALSN EETWKNRRRP RPIYSKDVLP ESLTQQNGST QNCSVTDGDL MVSAMPSLGL 300
301 KNPQELWGLT QNSLVFIEAL KLFFAKRKKE IGHLTFDKDD QLAVEFVTAA ANIRAESFGI 360
361 PLHSLFEAKG IAGNIVHAVA TTNAIIAGLI VIEAIKVLKK DVDKFRMTYC LEHPSKKLLL 420
421 MPIEPYEPNP ACYVCSETPL VLEINTRKSK LRDLVDKIVK TKLGMNLPLI MHGNSLLYEV 480
481 GDDLDDIMVA NYNANLEKYL SELPSPILNG SILTVEDLQQ ELSCKINVKH RFFSEILNPV 540
541 LNSVWFLIIL PSTFPKLFHF TESRNQDGLS LDIILGFSNV TIRRVLTMFE TGRRLTHPLL 600
601 ILFCHREEFD EEKEPEGMVL SGWTPSPATN GESASTSNNE NPVDVTESSS GSEPASKKRR 660
661 LSETEASNHK KETENVESED DDIMEVENPM MVSKKKIRVE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
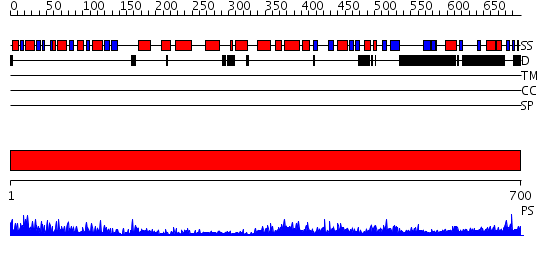
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..700] | 133.0 | SUMO E1 ACTIVATING ENZYME SAE1-SAE2-MG-ATP COMPLEX |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.62 |
Source: Reynolds et al. (2008)