













| General Information: |
|
| Name(s) found: |
GH35_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 612 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
auxin homeostasis
[TAS]
response to auxin stimulus [IMP][ISS] |
| Molecular Function: |
indole-3-acetic acid amido synthetase activity
[IDA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPEAPKKESL EVFDLTLDQK NKQKLQLIEE LTSNADQVQR QVLEEILTRN ADVEYLRRHD 60
61 LNGRTDRETF KNIMPVITYE DIEPEINRIA NGDKSPILSS KPISEFLTSS GTSGGERKLM 120
121 PTIEEELDRR SLLYSLLMPV MSQFVPGLEN GKGMYFLFIK SESKTPGGLP ARPVLTSYYK 180
181 SSHFKERPYD PYTNYTSPNE TILCSDSYQS MYSQMLCGLC QHQEVLRVGA VFASGFIRAI 240
241 KFLEKHWIEL VRDIRTGTLS SLITDPSVRE AVAKILKPSP KLADFVEFEC KKSSWQGIIT 300
301 RLWPNTKYVD VIVTGTMSQY IPTLDYYSNG LPLVCTMYAS SECYFGVNLR PLCKPSEVSY 360
361 TLIPSMAYFE FLPVHRNNGV TNSINLPKAL TEKEQQELVD LVDVKLGQEY ELVVTTYAGL 420
421 CRYRVGDLLR VTGFKNKAPQ FSFICRKNVV LSIDSDKTDE VELQNAVKNA VTHLVPFDAS 480
481 LSEYTSYADT SSIPGHYVLF WELCLDGNTP IPPSVFEDCC LAVEESFNTV YRQGRVSDKS 540
541 IGPLEIKIVE PGTFDKLMDY AISLGASINQ YKTPRCVKFA PIIELLNSRV VDSYFSPKCP 600
601 KWVPGHKQWG SN |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
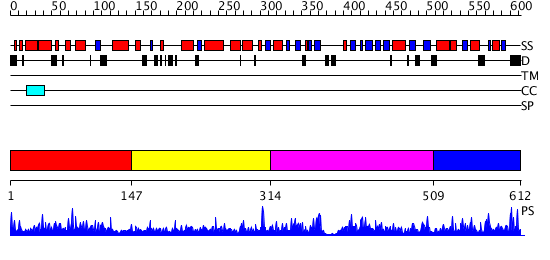
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..146] | 2.069975 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [147..313] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [314..508] | 1.075945 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [509..612] | 1.067986 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)