













| General Information: |
|
| Name(s) found: |
gi|15239528
[NCBI NR]
gi|10177266 [NCBI NR] |
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 596 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
intracellular signaling pathway
[IEA]
|
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTPDDFQLAT MPKLTVPFHD HPMSSSYCIF LESCSLCSFS KNSSWLRRSV TCISYECTDC 60
61 GLTLHKECVD HLFLNRPFLC NHVLKIYRNT FAWPDPDRKD DLSCHFCRRK LTSFFARLRN 120
121 LPSFFARCTI CNINVDKKCL MALGPPLAIF EPKHHEHSLT LLLRLVTFTC NACGVVDDRN 180
181 PYVCLECNLM VHKECVQNLP RVISINRHDH RISHTFHLGQ GEGDWECGVC RKTINWVYGG 240
241 YKCSRCPSYA VHSRCATRYE VWDGLELEDV PEEEEEIEDP FKVINEKGDI IHFSHEEHVL 300
301 RLDEIYVTDD ANMRCRCCVL AINGDPCYRC VECDFILHKA CANLPRKMRH LLHKHKLTLS 360
361 STGNRFLRCK ACDIKTNGFI YECLHEGCKS ISFTYDVRCS SVSEPFHHDL HQHPLYFTLE 420
421 KTNKCHACKR KIDWNPLNCT VCDDYSLCTR CAALPRKVKH RCDDHYLSLC QGVGSASGDL 480
481 WCDICETKTD PSVFYYTCDD CGLSLHIDCV LGDCYYTKVG CPGPRMEVHP NNGATRPICT 540
541 KCELRCKFPF FIKGTEEDLG IWYCSKDLCP KEFNFAVSPY RFALDIGLRD WDLRVF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
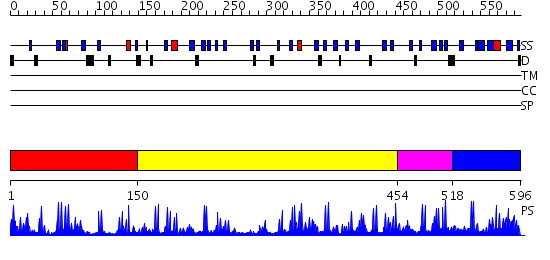
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..149] | 3.69897 | Crystal Structure of the Human Beta2-Chimaerin | |
| 2 | View Details | [150..453] | 0.987 | Crystal structure of N-terminal 3 domains of CI-MPR | |
| 3 | View Details | [454..517] | 4.154902 | Solution Structure of DC1 Domain of PDI-like Hypothetical Protein from Arabidopsis thaliana | |
| 4 | View Details | [518..596] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 | No functions predicted. | ||||||
| 3 |
|
||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)