













| General Information: |
|
| Name(s) found: |
SMC6B_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1057 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS]
chromosome [IEA] |
| Biological Process: |
sister chromatid cohesion
[IMP]
response to X-ray [IMP] double-strand break repair via homologous recombination [IMP] chromosome segregation [ISS] double-strand break repair [IMP] |
| Molecular Function: |
ATP binding
[IEA][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVKSGARASD SFIKQRSGSG SILRIKVENF MCHSNLQIEF GEWVNFITGQ NGSGKSAILT 60
61 ALCVAFGCRA RGTQRAATLK DFIKTGCSYA VVQVEMKNSG EDAFKPEIYG GVIIIERRIT 120
121 ESATATVLKD YLGKKVSNKR DELRELVEHF NIDVENPCVV MSQDKSREFL HSGNDKDKFK 180
181 FFFKATLLQQ VNDLLQSIYE HLTKATAIVD ELENTIKPIE KEISELRGKI KNMEQVEEIA 240
241 QRLQQLKKKL AWSWVYDVDR QLQEQTEKIV KLKERIPTCQ AKIDWELGKV ESLRDTLTKK 300
301 KAQVACLMDE STAMKREIES FHQSAKTAVR EKIALQEEFN HKCNYVQKIK DRVRRLERQV 360
361 GDINEQTMKN TQAEQSEIEE KLKYLEQEVE KVETLRSRLK EEENCFLEKA FEGRKKMEHI 420
421 EDMIKNHQKR QRFITSNIND LKKHQTNKVT AFGGDRVINL LQAIERNHRR FRKPPIGPIG 480
481 SHVTLVNGNK WASSVEQALG TLLNAFIVTD HKDSLTLRGC ANEANYRNLK IIIYDFSRPR 540
541 LNIPRHMVPQ TEHPTIFSVI DSDNPTVLNV LVDQSGVERQ VLAENYEEGK AVAFGKRLSN 600
601 LKEVYTLDGY KMFFRGPVQT TLPPLSRRPS RLCASFDDQI KDLEIEASKE QNEINQCMRR 660
661 KREAEENLEE LELKVRQLKK HRSQAEKVLT TKELEMHDLK NTVAAEIEAL PSSSVNELQR 720
721 EIMKDLEEID EKEAFLEKLQ NCLKEAELKA NKLTALFENM RESAKGEIDA FEEAENELKK 780
781 IEKDLQSAEA EKIHYENIMK NKVLPDIKNA EANYEELKNK RKESDQKASE ICPESEIESL 840
841 GPWDGSTPEQ LSAQITRMNQ RLHRENQQFS ESIDDLRMMY ESLERKIAKK RKSYQDHREK 900
901 LMACKNALDS RWAKFQRNAS LLRRQLTWQF NAHLGKKGIS GHIKVSYENK TLSIEVKMPQ 960
961 DATSNVVRDT KGLSGGERSF STLCFALALH EMTEAPFRAM DEFDVFMDAV SRKISLDALV1020
1021 DFAIGEGSQW MFITPHDISM VKSHERIKKQ QMAAPRS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
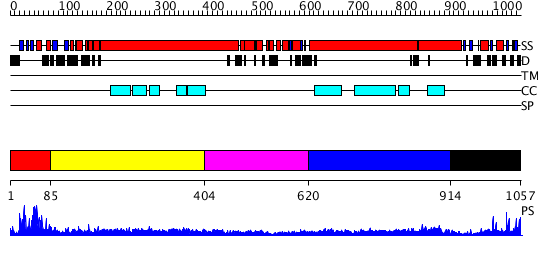
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..84] | 13.154902 | The Rad50 signature motif: essential to ATP binding and biological function | |
| 2 | View Details | [85..403] | 15.522879 | Heavy meromyosin subfragment | |
| 3 | View Details | [404..619] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [620..913] | 9.522879 | Tropomyosin | |
| 5 | View Details | [914..1057] | 17.154902 | Structural biochemistry of ATP-driven dimerization and DNA stimulated activation of SMC ATPases. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)