













| General Information: |
|
| Name(s) found: |
gi|15229491
[NCBI NR]
gi|11994636 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 662 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
intracellular signaling pathway
[IEA]
|
| Molecular Function: |
zinc ion binding
[IEA]
protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTFRLEGHEH RVSIVSSGDS LECDACDRSS GDGFSCSECK FTVHRKCAFV FMTEDIFHHP 60
61 SHDGHCLKLL TTGAPVHTDS KCHLCGKNTK HLLYHCYDCK VNLDIDCMID GICTLARLSK 120
121 PWHPHLLLMV DFSSDMWCDF CYEGGKHGYF CPRCRLVIHE SCVSVFDSPE INHPSHLSHP 180
181 LKLITNGAPD YTEDRSCHIC GKETGNLLYH CDICKFNLDM ECAVENPIPV ALSDLKVHEH 240
241 TLTLMPRLIS FICDACGTKG DRAPYVCVQC DFRMFHQKCA RLPRVIHVNH HDHRVSYKYP 300
301 LGPGEWRCGV CWEEIDWSYG AYSCSLCPNY AIHSLCATRK DVWDGKELDG VPEEVEDIEP 360
361 FKRNDDNTIT HFAHEHNLSL HNDDEESSLC GACVHPIGSY TFYKCSKSYC GFFLHERCAN 420
421 LATKKRHFLS PQPLDLSLQW IPYVLGRCRA CRQMFSEGFI YNSYRDKFDL LCSSITVPFI 480
481 HGSHDHHLLY YKLRYGQVKT CQNCGIDETE VLGCSKCNYY LDFCCATLPK TVMLPRYDDH 540
541 PLTLCYGDEN ASGKYWCDIC ERETNPKTWF YTCKDCGVTF HILCVVGDIR YAKPGGKING 600
601 DYELLANNSS SRPLCSTCHC RCPGPFILNK ERNNIFFCSY YCLGLSREHA YSSVDVCPPW 660
661 AI |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
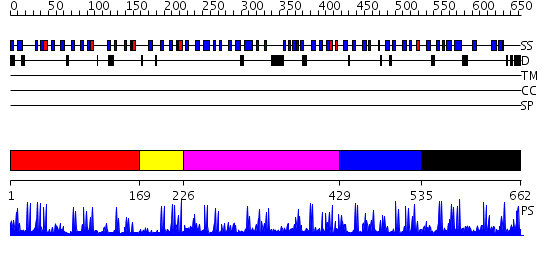
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..168] | 1.17 | ESCRT-II | |
| 2 | View Details | [169..225] | 6.09691 | NMR STRUCTURE OF A PROTEIN KINASE C-G PHORBOL-BINDING DOMAIN, MINIMIZED AVERAGE STRUCTURE | |
| 3 | View Details | [226..428] | 1.1 | Cysteine-rich (intestinal) protein, CRP, CRIP | |
| 4 | View Details | [429..534] | 4.045757 | No description for 2ysmA was found. | |
| 5 | View Details | [535..662] | 5.0 | Solution Structure of DC1 Domain of PDI-like Hypothetical Protein from Arabidopsis thaliana |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 |
|
||||||
| 3 |
|
||||||
| 4 | No functions predicted. | ||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)