













| General Information: |
|
| Name(s) found: |
PAH2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 930 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[IDA]
apoplast [IDA] |
| Biological Process: |
lipid metabolic process
[IGI]
cellular response to phosphate starvation [IGI] |
| Molecular Function: |
phosphatidate phosphatase activity
[IGI]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNAVGRIGSY IYRGVGTVSG PFHPFGGAID IIVVEQPDGT FKSSPWYVRF GKFQGVLKNG 60
61 RNLIRIDVNG VDSGFNMYLA HTGQAYFLRE VEDVVGESES GEVYTLSSGD EAETTSRDDV 120
121 VDKVKIPLKS RSCNYDSPSP RTGNGKIVGK PGILGYVFGG RSVRESQDCG VERAEIAADL 180
181 LEVKWSTNID TRKRGKGMSS ESLDGKDYGE STSTSGKSCV EGSSEMLVDS DSILETPLVA 240
241 SPTLRFLDEK EQDFRESTNV EDYCEEDGSS GVVVENGLCE ASSMVFSVTS EGSGNVEIFV 300
301 EPRTEALAED AVSGSDLDSK QELLRAPESV EIATLGSADQ ADMGSIGTSQ EGSSTGSPVQ 360
361 DENKITIKDM HISAGDFEKS QSASGESILQ PEIEEEQFSF SDLDECKPGG NSSVGSSSSD 420
421 TVKVDGKESY DETKTSPEKG VENTMALSEP INIERKKDIF TDEMERLVGS LPIMRLQNND 480
481 DMDASPSQPL SQSFDPCFNT SKLDLREDES SSGGLDAESV AESSPKLKAF KHVIANPEVV 540
541 ELSLCKHLLS EGMGAEAASQ AFNSEKLDME KFASLGPSIL ENDKLVVKIG GCYFPWDAAA 600
601 PIILGVVSFG TAQVFEPKGM IAVDRNEKPG DVLAQGSGSW KLWPFSLRRS TKEAEASPSG 660
661 DTAEPEEKQE KSSPRPMKKT VRALTPTSEQ LASLDLKDGM NSVTFTFSTN IVGTQQVDAR 720
721 IYLWKWNSRI VVSDVDGTIT RSDVLGQFMP LVGIDWSQTG VTHLFSAVKE NGYQLIFLSA 780
781 RAISQASVTR QFLVNLKQDG KALPDGPVVI SPDGLFPSLF REVIRRAPHE FKIACLEEIR 840
841 GLFPPEHNPF YAGFGNRDTD EISYLKVGIP RGKIFIINPK GEVAVNRRID TRSYTNLHTL 900
901 VNRMFPATSS SEPEDFNTWN FWKLPPPSLM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
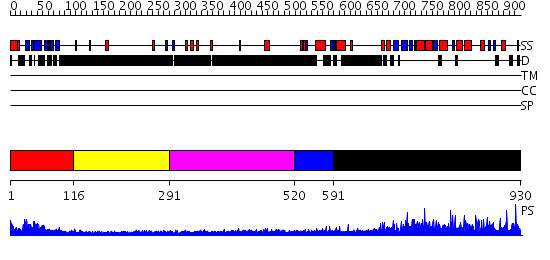
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..115] | 61.443697 | No description for PF04571.5 was found. No confident structure predictions are available. | |
| 2 | View Details | [116..290] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [291..519] | 1.371996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [520..590] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [591..930] | 1.27 | Polynucleotide kinase, phosphatase domain; Polynucleotide kinase, kinase domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)