













| General Information: |
|
| Name(s) found: |
IPT1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 357 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IEA]
|
| Biological Process: |
cytokinin biosynthetic process
[IGI][TAS]
secondary growth [IGI] |
| Molecular Function: |
adenylate dimethylallyltransferase activity
[IDA][TAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTELNFHLLP IISDRFTTTT TTSPSFSSHS SSSSSLLSFT KRRRKHQPLV SSIRMEQSRS 60
61 RNRKDKVVVI LGATGAGKSR LSVDLATRFP SEIINSDKIQ VYEGLEITTN QITLQDRRGV 120
121 PHHLLGVINP EHGELTAGEF RSAASNVVKE ITSRQKVPII AGGSNSFVHA LLAQRFDPKF 180
181 DPFSSGSCLI SSDLRYECCF IWVDVSETVL YEYLLRRVDE MMDSGMFEEL SRFYDPVKSG 240
241 LETRFGIRKA IGVPEFDGYF KEYPPEKKMI KWDALRKAAY DKAVDDIKRN TWTLAKRQVK 300
301 KIEMLKDAGW EIERVDATAS FKAVMMKSSS EKKWRENWEE QVLEPSVKIV KRHLVQN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
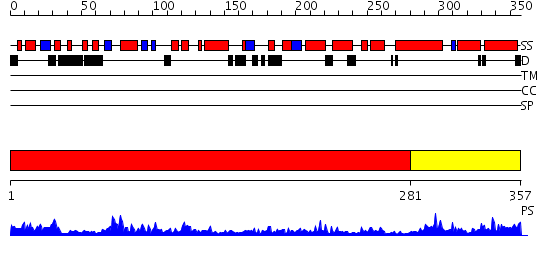
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..280] | 29.0 | HslU | |
| 2 | View Details | [281..357] | 51.69897 | No description for 2qgnA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)