













| General Information: |
|
| Name(s) found: |
UBA1C_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 613 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IEA]
|
| Biological Process: | NONE FOUND |
| Molecular Function: |
nucleic acid binding
[IEA]
nucleotide binding [IEA] zinc ion binding [IEA] RNA binding [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVWFQCDDCG ENLKKPRLPR HMSMCTATKF SCIDCGNMFG QVSVHYHNQC ITEAEKYGPM 60
61 VRSNGESSKQ KHDFDINAEL FNSQWFCSLC NATMTCEQDY FAHVYGKKHQ EKANEVADMD 120
121 YSKQQSEHPA VDKNNLTQQP DLDIYVGLSN DYPWFCSLCD INATSEQTLL AHANGKKHRV 180
181 KVERFDAEQQ KRQSTQHSTV DKKDYSKQQI EVDINVGLSN CYPWFCSLCN VKATCQQNLL 240
241 SHANGRKHRE NVELFDATQQ QQLEKTTVDK KDTTVNASDG NSEQKKVDLL VSSGVANGYS 300
301 QAHKKRKLET CDETWKREVV QAEEAKGGGE QKSESKKAKK QDKEKKRKKD KKQTKSDSDF 360
361 EHDKEDIKQL LVAYSKEELV NLIYKTAEKG SRLISAILES ADRDIAQRNI FVRGFGWDTT 420
421 QENLKTAFES YGEIEECSVV MDKDTGRGKG YGFVMFKTRK GAREALKRPE KRMYNRIVVC 480
481 NLASEKPGKA GKEQDMAEPV NIDLTKMANQ SEAVLPGIEL GRGHVLEKMH HQQQQTMDMF 540
541 GQNMPFYGYS HQFPGFDPMY GALSGNQMLA GLPNYGMFGS GMMTNQGSML PPPNHLGMAG 600
601 QYFGDGEQAW HQR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
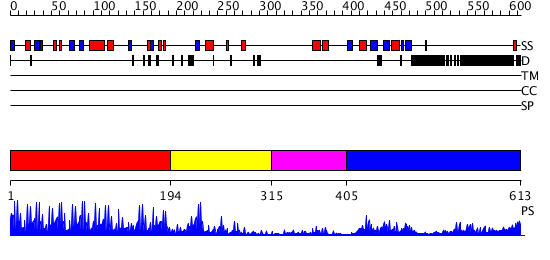
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..193] | 53.30103 | No description for 2i13A was found. | |
| 2 | View Details | [194..314] | 2.22 | Solution Structure of the N-terminal Zinc Fingers of the Xenopus laevis double stranded RNA binding protein ZFa | |
| 3 | View Details | [315..404] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [405..613] | 23.69897 | Crystal Structure of UP1 Complexed With d(TTAGGGTTAG(6-MI)G); A Human Telomeric Repeat Containing 6-methyl-8-(2-deoxy-beta-ribofuranosyl)isoxanthopteridine (6-MI) |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)