













| General Information: |
|
| Name(s) found: |
CTF64_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 461 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
RNA 3'-end processing
[IMP]
developmental growth [IMP] |
| Molecular Function: |
RNA binding
[ISS]
protein binding [IPI] mRNA binding [IDA] transcription repressor activity [IMP] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASSSSQRRC VFVGNIPYDA TEEQLREICG EVGPVVSFRL VTDRETGKPK GYGFCEYKDE 60
61 ETALSARRNL QSYEINGRQL RVDFAENDKG TDKTRDQSQG GPGLPSTTTV TESQKQIGGP 120
121 VDSNMHQPVG LHLATTAASV IAGALGGPQV GSQFTQSNLQ VPASDPLALH LAKMSRSQLT 180
181 EIISSIKLMA TQNKEHARQL LVSRPQLLKA VFLAQVMLGI VSPQVLQSPN IVQAPSHMTG 240
241 SSIQDAQLSG QNLLPPLAQR SQQLSRAPHS QYPVQQSSKQ PFSQIPQLVA QPGPSSVNPP 300
301 PRSQVKVENA PFQRQQVVPA STNIGYSSQN SVPNNAIQPS QVPHQALPNS VMQQGGQTVS 360
361 LNFGKRINEG PPHQSMNRPS KMMKVEDRRT TSLPGGHVSN SMLPNQAQAP QTHISPDVQS 420
421 TLLQQVMNLT PEQLRLLTPE QQQEVLKLQQ ALKQDHMMQP S |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
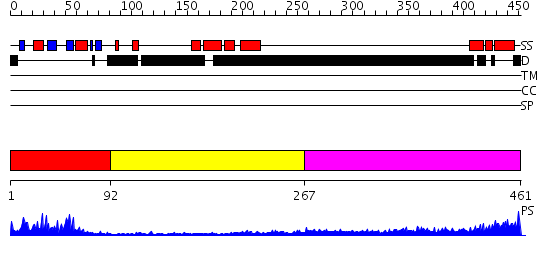
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..91] | 24.0 | NMR Structure of the N-terminal RRM domain of Cleavage stimulation factor 64 KDa subunit | |
| 2 | View Details | [92..266] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [267..461] | 11.0 | poly(A) binding protein |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 |
|
|||||||||||||||
| 2 | No functions predicted. | |||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.55 |
Source: Reynolds et al. (2008)