













| General Information: |
|
| Name(s) found: |
gi|7484877
[NCBI NR]
gi|7269597 [NCBI NR] gi|4972068 [NCBI NR] gi|3327868 [NCBI NR] gi|186514102 [NCBI NR] gi|15234126 [NCBI NR] |
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1058 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
plasma membrane [IDA] |
| Biological Process: |
anthocyanin biosynthetic process
[IDA]
chlorophyll biosynthetic process [IDA] response to light stimulus [IEP] |
| Molecular Function: |
transcription activator activity
[IGI]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDPRTRLDYA LFQLTPTRTR CDLVIFSGGE NEKLASGIFQ PFVTHLKSVS DQISKGGYSV 60
61 TLRPSSVGVP WFTKVTLQRF VRFVTTPEVL ERSVTLEKEI EQIEDSIQAN AAAIAGEAEG 120
121 NELGGTWTSQ KSTALSKTKG ETDGDTVEEN SKVGLQRVLE NRKAALCKEQ AMAYARALVV 180
181 GFELDYMDDL FSFADAFGAS RLREACVNFV DLCKRKNEDR MWVDQITAMQ AFPRPELTFM 240
241 GDSGIVLAGE ENDLLNATNV KHGNSMDASS QGSFETGQEG RAQMAMPWPN QFPQYMQNFQ 300
301 GHGYPPPYMF PGMQGQSPYF HGNMQWPVNM GDVESNEKSS KKKKKKKKNK KKSKQDESAE 360
361 PSDNSSTETE SEDGNEGKKQ SRKVVIRNIN YITSKRNGAK ESDSDESGEE EGFVDGDSIK 420
421 QQVEEAIGSV ERRHKSTSHR QRKHKSHNGD DDSSNKETKG NDNWDAFQNL LLKDNDSEPE 480
481 ELLRISSTAL NMASEVVRKR EPPSDDSFLV AIGNEDWGRE TSIEKFNAGE NVRIIRKGNN 540
541 YDEEMLNPGR SDESRSYSQA EMSVHDGKLR TRNEAEEDWF IRNQAGPETD PSLVKTFVGD 600
601 HFHLNKSSER DVLTDDSFMI HSRVENQVED SRLRTEIMDL DVYGTTQQEN SAPENTPHEP 660
661 DDLYMVLGRE QDVKPTLLPW TPEIDFETNT LAQRTSRIDL ITATKASAGE QTLDGKEKKS 720
721 RGISKGKDAK SRASSRPDPA SKAKRPAWGS RAAVSKSKSE MEEERKKRME ELLIQRQKRI 780
781 AEKSSGGSVS SSLASKKTPT VTKSVKSSIK NEKTPEAAQS KAKPVLRSST IERLAVARTA 840
841 PKEPQQKPVI KRTSKPSGYK TEKAQEKKSS KIGQSDAKSV ELSRDPSLEI KETVVEDSHS 900
901 YLSEKQVDAL PAVASVDDFK DIKELHSLPS EETARVKNRP NEIIAEKVQD QTKIDDQETV 960
961 KNTSVSEDKQ ITTKHYSEDV GEVQASQEKP VSPKKSVTFS ETNMEEKYYF SPAVSEIDIS1020
1021 TPPATEADHS RKKWNSEETS PKATAKVFRK LLMFGRKK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
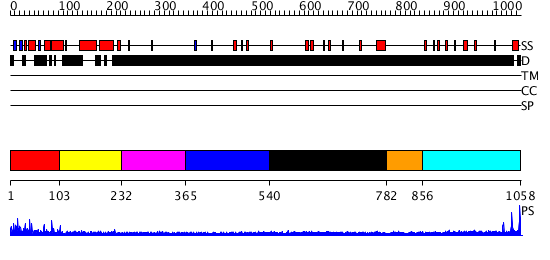
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..102] | 1.20798 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [103..231] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [232..364] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [365..539] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [540..781] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [782..855] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [856..1058] | 2.542997 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)