













| General Information: |
|
| Name(s) found: |
ZW10_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 742 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
chromosome, centromeric region [IEA] |
| Biological Process: |
chromosome segregation
[ISS]
|
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPEIDALFES INVRDLLAGH DLNDPTTPLS APDLRLLINR LESHSLRIKS KVQSYLVAHH 60
61 SDFSELFSLC QDTVSRTRLI SDDVSDVLQL VSDRPIDVEI RSVVDEITEK TKEVKLKRES 120
121 LDLVNAIVGI CEALQETKEA LKNGRFRFAA ERIRELKVVL RIGEEEDGEP VAYALLRKEW 180
181 SNCFDEIQEV LAKFMENAVR FELDSSRIRI KYQLSVGETA GIALSTVLEA MEVIGILDYG 240
241 LAKAADSIFK HVITPAVTHA STFAAVEDLC KSAGEVTEAT LRLEQSSDHK FEDVDGDAMY 300
301 SGILKVVKFI CSSLCFGNVT WIHSFGRLTW PRISELIISK FLSKVVPEDA SKLADFQKII 360
361 ERTSQFEAAL KELNFVSSSD AESRLSKYAE DVEVHFASRK KIEILAKARN LLLQCNFTIP 420
421 QDIAMRNAKH IVCLLFSSER CVVSEAASQL MNLVHKTLED VCVSSARVAS EFYNAARDSI 480
481 LLYEAVVPVK LEKQLDGLNE AAVLLHNDCL YLFEEILGLA FEYRASFPSS IKEYAVFADI 540
541 APRFKLMAEE VLQKQVHLVI SSLREAIDSA DGFQNTHQIK QFKSAEFSID QVVFSLKNVH 600
601 MIWEPVLRPK TYKQSMCAVL ESVFRRIARD ILLLDDMAAD ETFELQKLIY LMLKNLSSVL 660
661 DSVRSADETS RPLDDIIPSL RKTRKLAELL DMPLMSITSA WESGELFRCN FTRTEVQDFI 720
721 KAIFTDSPLR KECLWRIDEV NQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
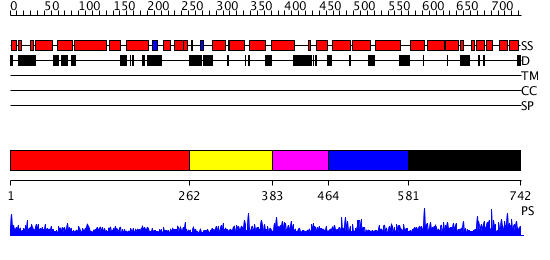
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..261] | 3.69897 | Heavy meromyosin subfragment | |
| 2 | View Details | [262..382] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [383..463] | 1.051995 | View MSA. Confident ab initio structure predictions are available. | |
| 4 | View Details | [464..580] | N/A | Confident ab initio structure predictions are available. | |
| 5 | View Details | [581..742] | 1.037982 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)