













| General Information: |
|
| Name(s) found: |
YU20_ARATH
[Swiss-Prot]
NOC2L_ARATH [Swiss-Prot] |
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 779 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGAKELKGFE IDKHFKSNVD DKKRVKKLKS KKLEAEEELN NVQEIDAHDI VMEQKSDKKR 60
61 GKKVKSKKAE AEEHEEELKR LQEKDPDFFQ YMKEHDAELL KFDATEIEDD ADVEPDTDLE 120
121 DTEKEGDDEA TKMEIAKKVH VQKTITASMV DAWSKSIEDE AKLGGVRSIL RAYRTACHYG 180
181 DDTGDDQSTK FSVMSSEVFN KIMIYVLSEM DGILRKLLRF PEDTRGTKET ILELTNTRPW 240
241 KNYNHLVKSY LGNSLHVLNQ MTDTEMITFT LRRLKHSSVF LAAFPSLLRK YIKVALHFWG 300
301 TGSGALPVVS LLFLRDLCIR LGSDCVDDCF KGMYKAYVLN CQFVNADKLK HISFLGNCFI 360
361 ELLGTDISAA YQHAFVFIRQ LAMILREALN TKTKEAFRKV YQWKFIHCLE LWTGAVCAYS 420
421 SQSELRPVAY PLAQIITGVA RLVPTARYTP LRLRCVRMLN RLAAATGTFI PVSMLLVDML 480
481 EMKELNRPPT GGVGKGVDLR TLLKVSKPAV KTRAFQEACV YTVVEELVEH LSQWSCSVAF 540
541 FELSFIPTIR LRSFCKSTKA ERFRKEMKQL ISQIEANSEF VNKKRALIKF LPNDLAAESF 600
601 LEDEKKAGKT PLLQYAEIIR QRAQQRNESL VESDVIVGEN SAVFGKNAPS SDDEDDEDRM 660
661 EKGAAAFNSS WLPGSDSKEK EPEEEKTKKK KRKRGGKSKT EKKQDEQGLG EDDVVEDFVL 720
721 SSDEEEEDLF DIGGDKDEDD AVDEIADPET KTSKKTKGTY KTWHKAYKKT KKKKARVAS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
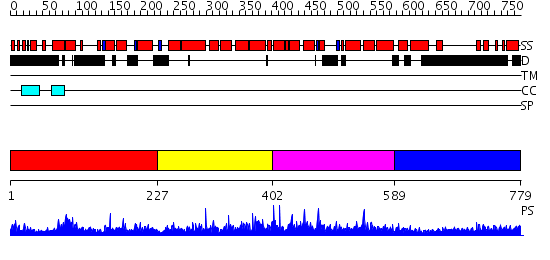
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..226] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [227..401] | 1.053953 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [402..588] | 1.078961 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [589..779] | 18.095997 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.40 |
Source: Reynolds et al. (2008)